ankyrin repeat and SOCS box containing 2b
ZFIN 
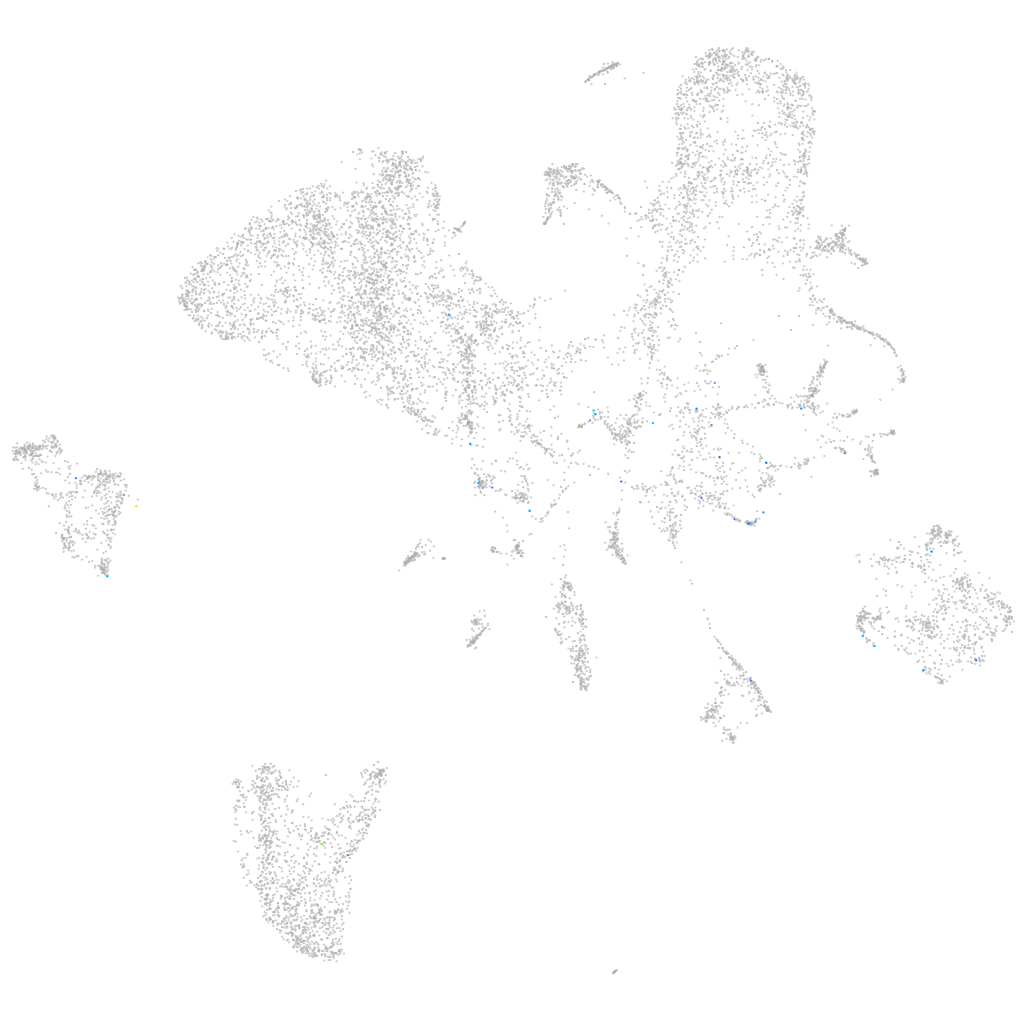
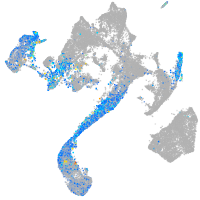
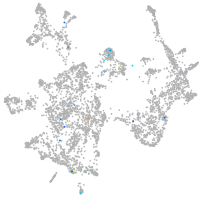
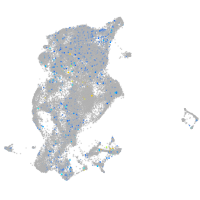
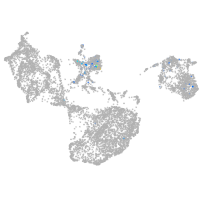
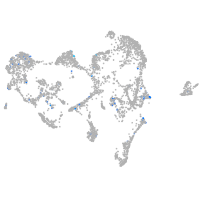
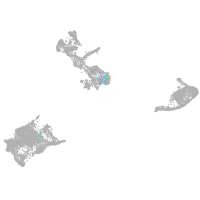
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| XLOC-030940 | 0.369 | aldob | -0.056 |
| FO834828.1 | 0.356 | gapdh | -0.055 |
| zgc:153118 | 0.315 | gamt | -0.051 |
| BX539313.1 | 0.306 | atp5fa1 | -0.049 |
| alpk3a | 0.273 | eno3 | -0.049 |
| CR388040.1 | 0.260 | ahcy | -0.048 |
| xkr6a | 0.249 | sod1 | -0.047 |
| CT009714.1 | 0.224 | acadm | -0.047 |
| stac3 | 0.222 | aldh6a1 | -0.047 |
| edil3b | 0.222 | gpx4a | -0.043 |
| LOC103911600 | 0.217 | COX5B | -0.043 |
| CU019663.1 | 0.211 | glud1b | -0.043 |
| CABZ01039863.1 | 0.209 | atp5f1b | -0.042 |
| LOC103909593 | 0.208 | nupr1b | -0.042 |
| oprd1a | 0.208 | atp5mc3b | -0.042 |
| si:ch211-225p5.8 | 0.201 | gstt1a | -0.042 |
| aqp4 | 0.194 | cx32.3 | -0.042 |
| eya4 | 0.187 | gstk1 | -0.041 |
| ppp1r9ba | 0.183 | fbp1b | -0.041 |
| prrt2 | 0.181 | suclg2 | -0.041 |
| kif3cb | 0.178 | bhmt | -0.040 |
| TTC28 | 0.175 | uqcrfs1 | -0.040 |
| BX649485.2 | 0.174 | sod2 | -0.040 |
| CR769778.1 | 0.171 | mt-nd2 | -0.040 |
| ahdc1 | 0.171 | scp2a | -0.040 |
| kif26aa | 0.171 | gatm | -0.040 |
| actc1c | 0.169 | dap | -0.040 |
| nr4a2b | 0.168 | pgm1 | -0.040 |
| parp6b | 0.167 | cat | -0.039 |
| vopp1 | 0.166 | atp5pd | -0.039 |
| XLOC-021387 | 0.158 | pklr | -0.039 |
| nkain2 | 0.155 | gstp1 | -0.039 |
| shox | 0.154 | agxta | -0.039 |
| dixdc1a | 0.153 | nipsnap3a | -0.039 |
| ugt1b3 | 0.152 | suclg1 | -0.039 |