ADP-ribosylation factor-like 6 interacting protein 6
ZFIN 
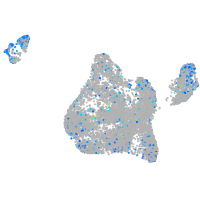
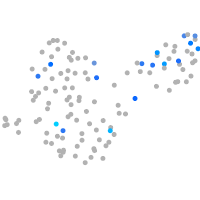
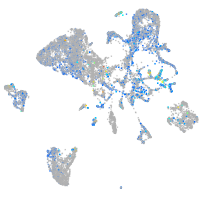
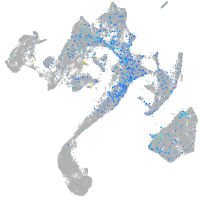
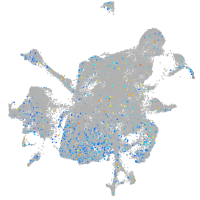
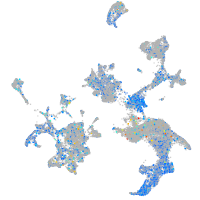
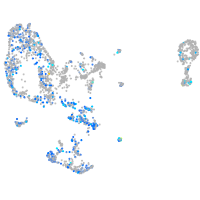
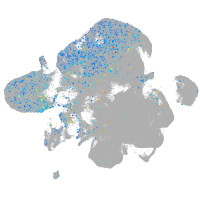
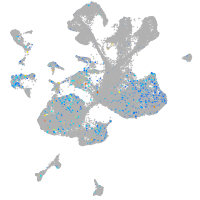
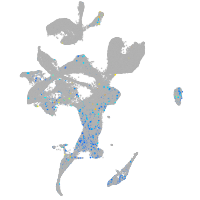
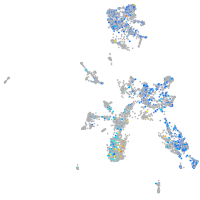
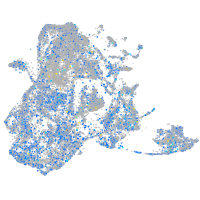
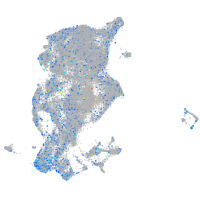
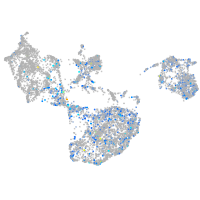
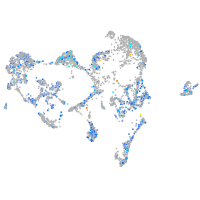
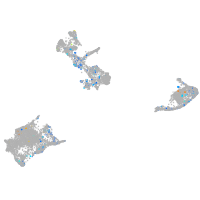
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| scg3 | 0.311 | gamt | -0.172 |
| vat1 | 0.267 | bhmt | -0.159 |
| rem2 | 0.260 | gatm | -0.157 |
| neurod1 | 0.259 | apoc1 | -0.155 |
| pcsk1nl | 0.258 | ahcy | -0.136 |
| insm1b | 0.253 | gapdh | -0.135 |
| c2cd4a | 0.251 | pnp4b | -0.133 |
| scgn | 0.246 | agxtb | -0.132 |
| fev | 0.243 | apoa1b | -0.126 |
| zgc:101731 | 0.241 | mat1a | -0.123 |
| si:ch73-160i9.2 | 0.240 | fetub | -0.123 |
| calm1b | 0.238 | apoa2 | -0.122 |
| gfra1a | 0.235 | afp4 | -0.122 |
| mir7a-1 | 0.233 | aqp12 | -0.121 |
| arxa | 0.231 | rbp4 | -0.119 |
| rasd4 | 0.230 | serpina1 | -0.116 |
| anxa4 | 0.229 | fabp10a | -0.115 |
| hepacam2 | 0.227 | rbp2b | -0.115 |
| egr4 | 0.226 | zgc:123103 | -0.114 |
| rprmb | 0.226 | hao1 | -0.114 |
| pcsk2 | 0.221 | tfa | -0.113 |
| tango2 | 0.220 | ces2 | -0.113 |
| si:ch1073-429i10.3.1 | 0.220 | apoa4b.1 | -0.113 |
| pax6b | 0.218 | apom | -0.111 |
| elovl1b | 0.217 | LOC110437731 | -0.110 |
| calm3b | 0.217 | serpina1l | -0.108 |
| eml2 | 0.216 | apobb.1 | -0.106 |
| capsla | 0.216 | ttr | -0.106 |
| h3f3a | 0.215 | ambp | -0.105 |
| gapdhs | 0.212 | pck1 | -0.104 |
| elovl1a | 0.211 | gc | -0.104 |
| pcsk1 | 0.210 | uox | -0.104 |
| cacna1c | 0.210 | si:dkey-86l18.10 | -0.104 |
| ccka | 0.209 | fgg | -0.103 |
| nkx2.2a | 0.208 | comtd1 | -0.103 |