ADP-ribosylation factor-like 6 interacting protein 5a
ZFIN 
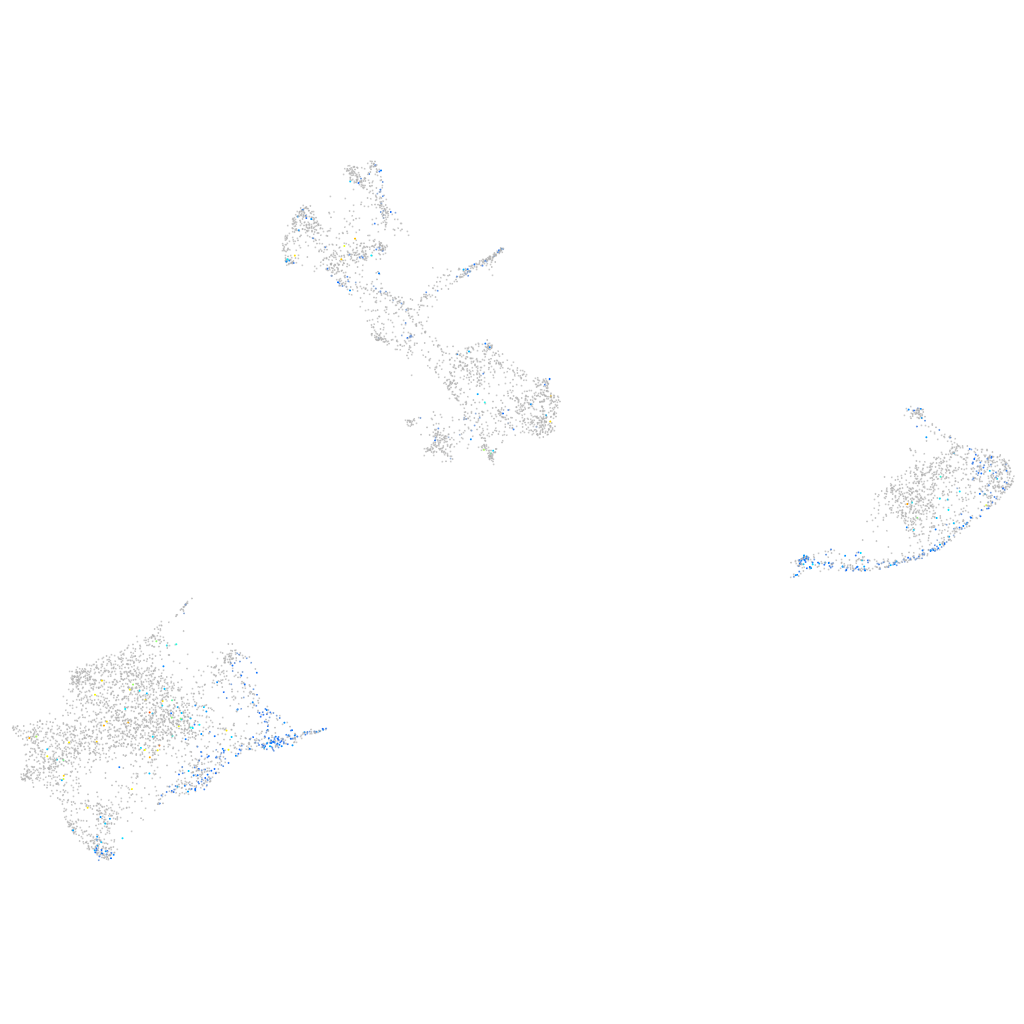
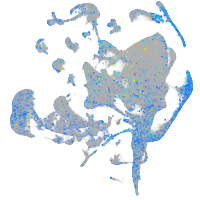
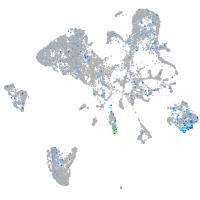
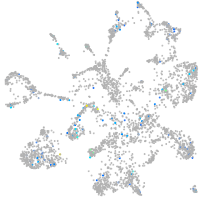
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| xbp1 | 0.133 | pvalb1 | -0.093 |
| crestin | 0.131 | pvalb2 | -0.084 |
| mitfa | 0.128 | hbae3 | -0.075 |
| ctsc | 0.128 | actc1b | -0.074 |
| ctsba | 0.127 | hbbe1.3 | -0.070 |
| cx30.3 | 0.125 | ptmaa | -0.068 |
| gtpbp3 | 0.123 | tuba1c | -0.061 |
| si:dkey-56m15.5 | 0.122 | hbae1.1 | -0.061 |
| lman2 | 0.120 | tmsb | -0.055 |
| rgs2 | 0.118 | elavl3 | -0.055 |
| msx1b | 0.116 | hbbe1.1 | -0.054 |
| sigmar1 | 0.115 | nova2 | -0.053 |
| pcdh9 | 0.115 | sncb | -0.053 |
| scpep1 | 0.109 | tnni2a.4 | -0.053 |
| ssr3 | 0.109 | mdkb | -0.052 |
| atf3 | 0.108 | myhz1.1 | -0.051 |
| ednrab | 0.108 | vamp2 | -0.050 |
| gphna | 0.108 | gpm6aa | -0.050 |
| tram1 | 0.108 | stmn2a | -0.048 |
| XLOC-001964 | 0.108 | stx1b | -0.048 |
| ssr2 | 0.108 | hbbe1.2 | -0.047 |
| ssr4 | 0.107 | hbae1.3 | -0.046 |
| dad1 | 0.107 | hmgb3a | -0.045 |
| fkbp2 | 0.107 | tpma | -0.045 |
| slc15a2 | 0.107 | CU467822.1 | -0.045 |
| aamp | 0.106 | dpysl3 | -0.045 |
| canx | 0.106 | ckma | -0.045 |
| rnaseka | 0.106 | zgc:110339 | -0.044 |
| rabl6b | 0.106 | nme2b.2 | -0.044 |
| pim1 | 0.105 | apoa1b | -0.044 |
| C12orf75 | 0.105 | cdh2 | -0.044 |
| tpp1 | 0.104 | nsg2 | -0.044 |
| npm1a | 0.104 | hmgb1b | -0.043 |
| rab32a | 0.104 | krt4 | -0.042 |
| slc1a3a | 0.104 | mylz3 | -0.042 |