ADP-ribosylation factor-like 14
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
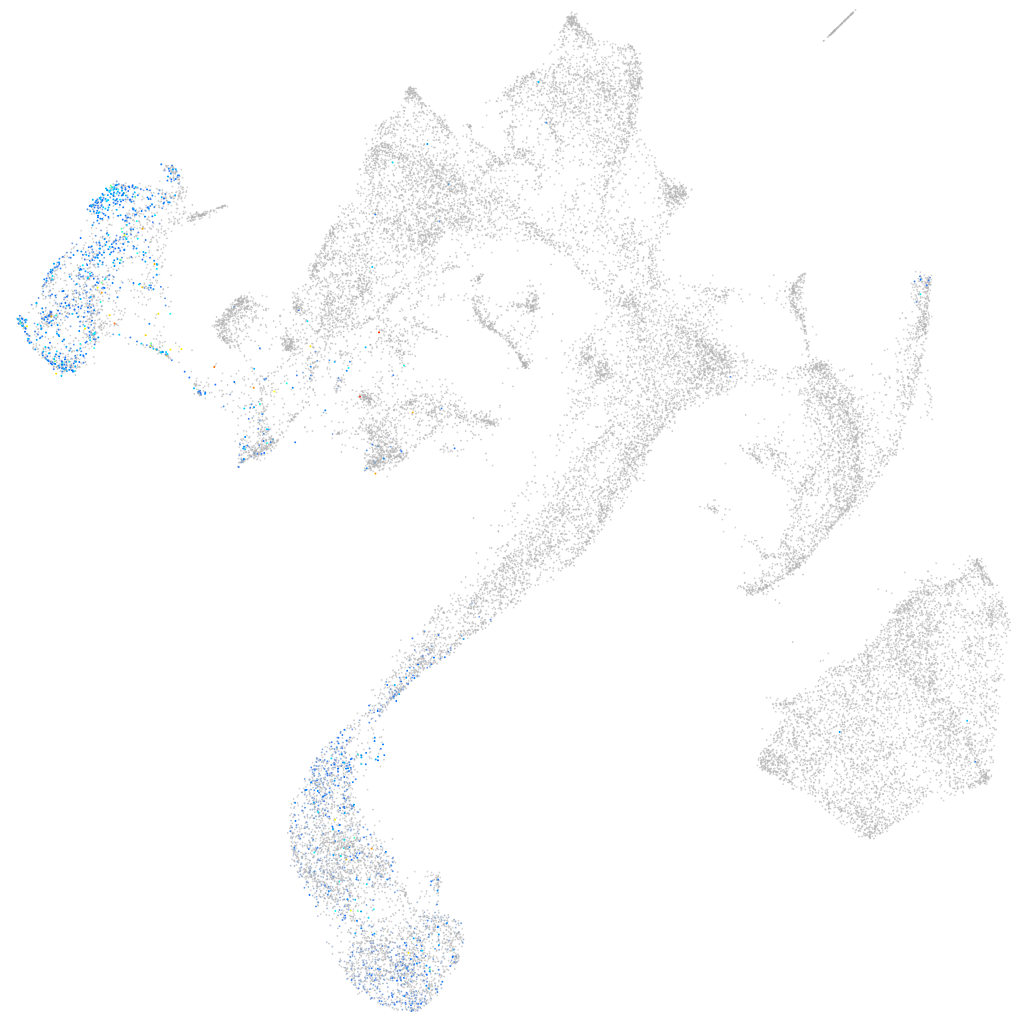
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| tnnc1b | 0.407 | hsp90ab1 | -0.317 |
| myom1b | 0.391 | pabpc1a | -0.292 |
| ryr1a | 0.389 | h3f3d | -0.285 |
| LO018250.1 | 0.384 | h2afvb | -0.282 |
| tnni4b.2 | 0.381 | ran | -0.282 |
| hspb11 | 0.377 | khdrbs1a | -0.279 |
| pgam2 | 0.375 | si:ch73-1a9.3 | -0.276 |
| xirp2a | 0.375 | hmgb2b | -0.274 |
| myoz2b | 0.374 | hnrnpaba | -0.270 |
| smyhc1 | 0.374 | cirbpb | -0.266 |
| casq2 | 0.373 | hnrnpabb | -0.260 |
| nrap | 0.372 | si:ch211-222l21.1 | -0.254 |
| tcap | 0.371 | hmgn2 | -0.252 |
| eno3 | 0.368 | setb | -0.249 |
| mustn1b | 0.367 | hnrnpa0b | -0.247 |
| tnni1c | 0.365 | hmgb2a | -0.247 |
| myl13 | 0.364 | cirbpa | -0.246 |
| hhatlb | 0.364 | snrpf | -0.246 |
| cavin4b | 0.363 | hmga1a | -0.245 |
| srl | 0.362 | ubc | -0.243 |
| tpm2 | 0.361 | ptmab | -0.243 |
| si:ch211-266g18.10 | 0.360 | si:ch73-281n10.2 | -0.242 |
| prx | 0.357 | cfl1 | -0.241 |
| gapdh | 0.357 | cbx3a | -0.240 |
| klhl43 | 0.348 | hmgn7 | -0.238 |
| neb | 0.348 | syncrip | -0.238 |
| aldoab | 0.348 | naca | -0.236 |
| pvalb4 | 0.347 | eef1a1l1 | -0.236 |
| si:ch211-28p3.4 | 0.346 | actb2 | -0.235 |
| atp2a1 | 0.346 | snrpd1 | -0.234 |
| CU633479.2 | 0.344 | snrpb | -0.230 |
| smyd1a | 0.343 | tuba8l4 | -0.230 |
| nme2b.2 | 0.341 | h2afva | -0.227 |
| cavin4a | 0.341 | snrpd2 | -0.227 |
| trdn | 0.341 | eif5a2 | -0.225 |