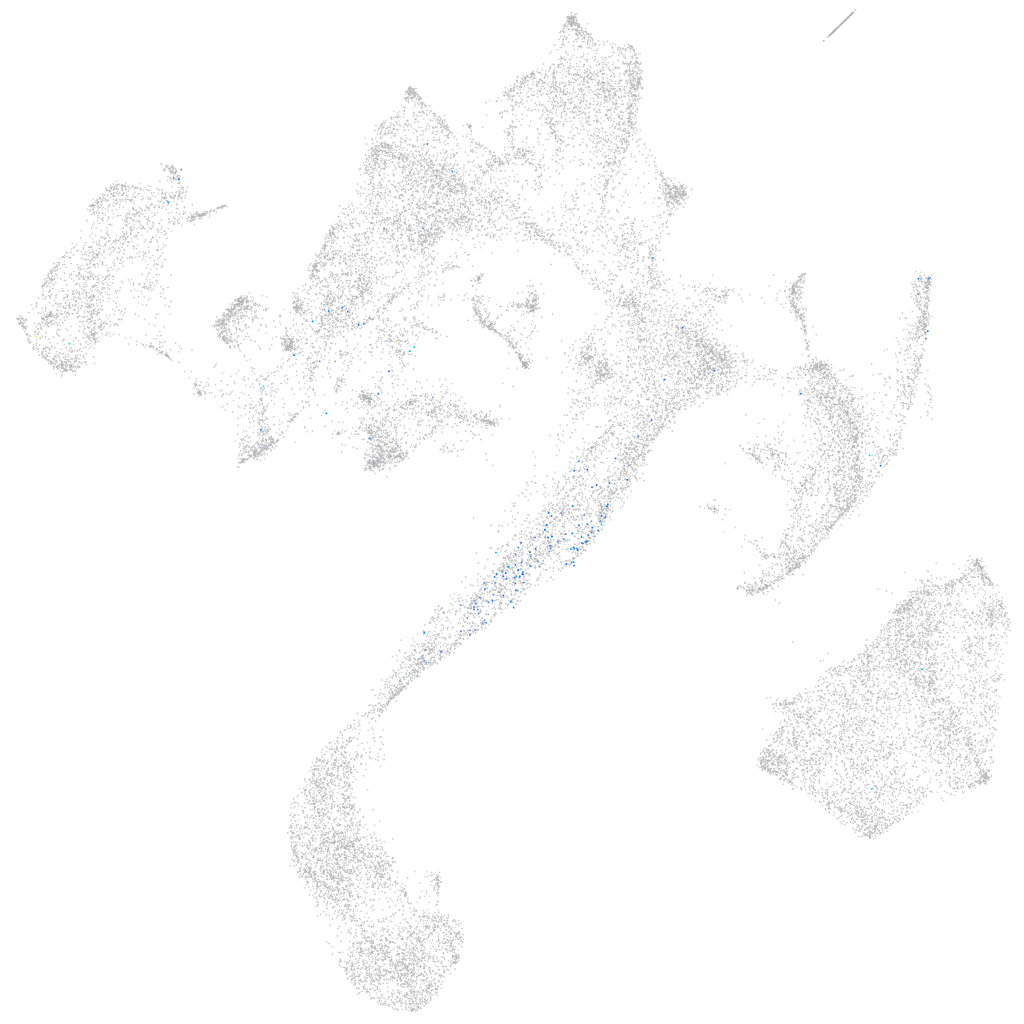
Rho guanine nucleotide exchange factor (GEF) 3
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| CU984583.1 | 0.163 | ptmab | -0.060 |
| mymk | 0.157 | si:ch211-222l21.1 | -0.057 |
| sh3d19 | 0.154 | tmsb4x | -0.050 |
| efhd1 | 0.148 | hmgb1b | -0.049 |
| si:dkeyp-19e1.3 | 0.146 | tuba8l4 | -0.044 |
| si:ch1073-268j14.1 | 0.145 | tubb2b | -0.043 |
| hjv | 0.145 | cbx5 | -0.042 |
| s1pr5a | 0.145 | stmn1a | -0.042 |
| mafbb | 0.141 | hmgb2a | -0.041 |
| myog | 0.137 | wls | -0.041 |
| CR381686.5 | 0.136 | fxyd6l | -0.041 |
| rnf41l | 0.135 | si:ch211-288g17.3 | -0.040 |
| aktip | 0.134 | tubb4b | -0.040 |
| fbxl22 | 0.133 | si:dkey-56m19.5 | -0.039 |
| traf4a | 0.131 | tcf15 | -0.039 |
| mafaa | 0.131 | hmgb2b | -0.039 |
| flncb | 0.131 | anp32b | -0.038 |
| foxd3 | 0.130 | pcna | -0.038 |
| filip1a | 0.130 | ctsla | -0.038 |
| si:ch211-131k2.3 | 0.125 | hmgb1a | -0.037 |
| mss51 | 0.123 | chaf1a | -0.037 |
| tmx2a | 0.122 | xbp1 | -0.036 |
| crip2 | 0.122 | map2k6 | -0.036 |
| rgs18 | 0.120 | sumo3b | -0.036 |
| zgc:101853 | 0.120 | dynll1 | -0.036 |
| znf385a | 0.120 | pmp22a | -0.036 |
| mef2d | 0.120 | mcm6 | -0.036 |
| fam107b | 0.120 | si:ch73-1a9.3 | -0.036 |
| ptprua | 0.119 | dek | -0.036 |
| reep1 | 0.119 | anp32a | -0.035 |
| jam2a | 0.119 | fkbp7 | -0.035 |
| nexn | 0.119 | sumo3a | -0.035 |
| cygb1 | 0.116 | hmgn7 | -0.035 |
| phactr3b | 0.114 | CABZ01080702.1 | -0.035 |
| rbfox1l | 0.113 | mcm7 | -0.035 |