Rho GTPase activating protein 31
ZFIN 
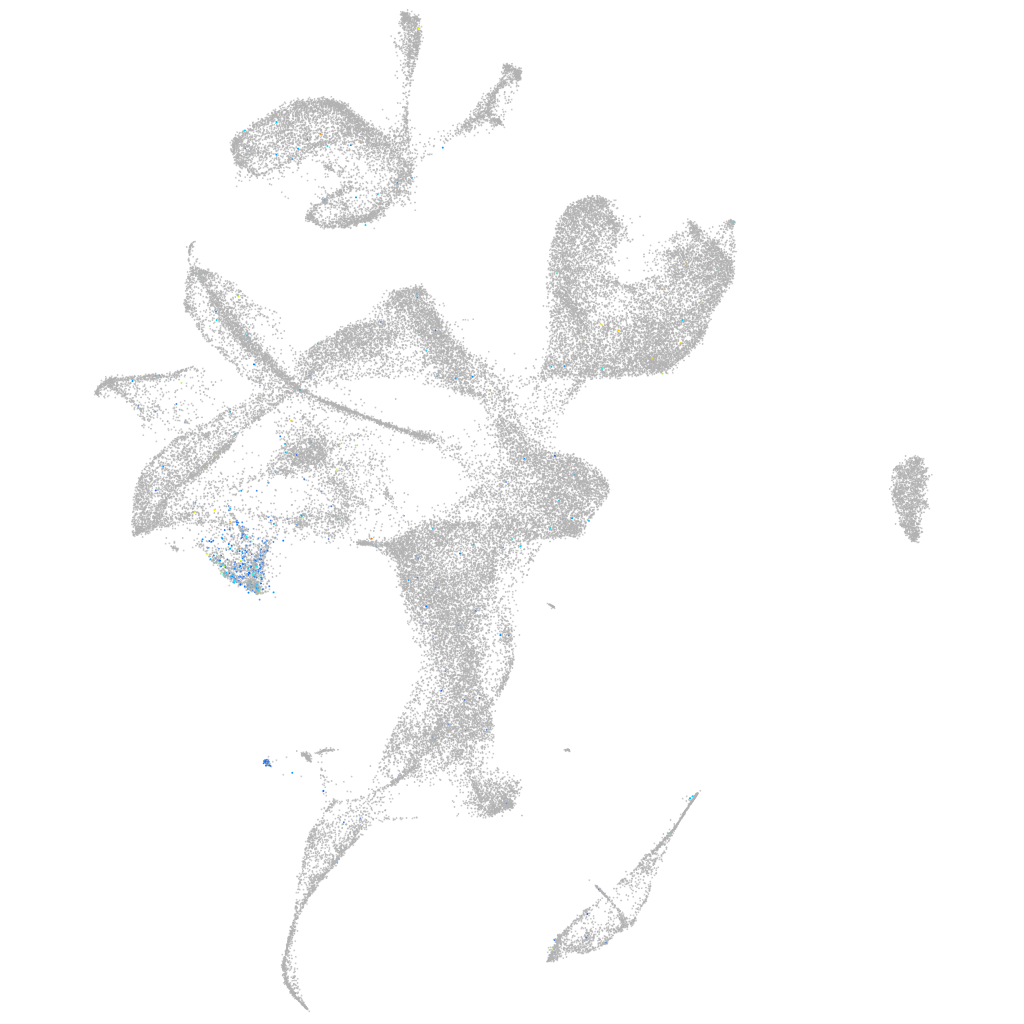
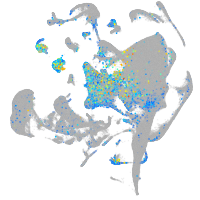
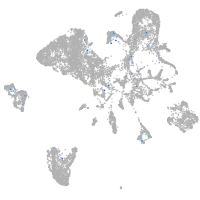
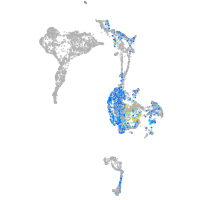
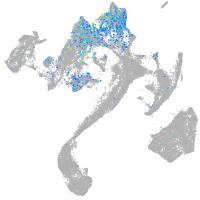
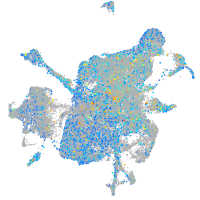
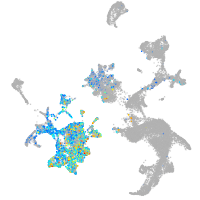
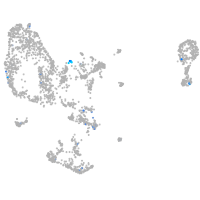
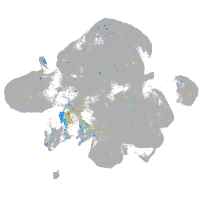
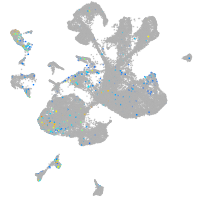
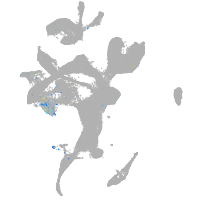
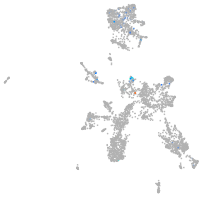
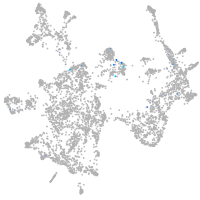
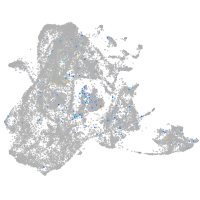
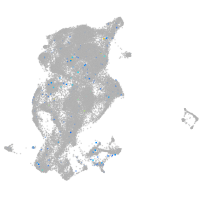
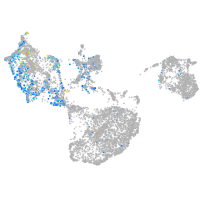
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| pmp22a | 0.209 | pax6a | -0.025 |
| krt18b | 0.174 | rrm1 | -0.024 |
| col5a1 | 0.165 | ptmab | -0.024 |
| col4a1 | 0.161 | slbp | -0.023 |
| CU929237.1 | 0.159 | CABZ01005379.1 | -0.022 |
| si:dkey-261h17.1 | 0.158 | pax6b | -0.021 |
| twist1a | 0.156 | sinhcafl | -0.021 |
| ms4a17a.9 | 0.149 | seta | -0.020 |
| col6a2 | 0.146 | hes6 | -0.020 |
| krt8 | 0.143 | cadm3 | -0.020 |
| col1a2 | 0.140 | rorb | -0.020 |
| pdgfra | 0.139 | ccna2 | -0.020 |
| dcn | 0.138 | zgc:153405 | -0.019 |
| fmoda | 0.138 | pcna | -0.019 |
| si:ch1073-291c23.2 | 0.138 | krcp | -0.019 |
| col5a2a | 0.137 | chaf1a | -0.019 |
| mfap2 | 0.135 | hmga1a | -0.019 |
| SCARF2 | 0.135 | si:ch211-137a8.4 | -0.019 |
| ecrg4a | 0.135 | lig1 | -0.019 |
| mxra8b | 0.134 | neurod4 | -0.019 |
| col11a1b | 0.134 | mibp | -0.019 |
| lrrc17 | 0.134 | zgc:110216 | -0.019 |
| tuba8l3 | 0.134 | khdrbs1a | -0.019 |
| twist2 | 0.131 | zgc:110425 | -0.018 |
| LO018188.1 | 0.130 | XLOC-042899 | -0.018 |
| ahnak | 0.130 | si:dkey-261m9.17 | -0.018 |
| mmp14a | 0.128 | hes2.2 | -0.018 |
| entpd1 | 0.127 | ccne2 | -0.018 |
| fibina | 0.127 | zic2b | -0.018 |
| barx1 | 0.127 | si:ch211-114n24.6 | -0.018 |
| col6a3 | 0.127 | prim2 | -0.018 |
| twist1b | 0.126 | lbr | -0.018 |
| col6a1 | 0.126 | tubb2b | -0.017 |
| tmem176 | 0.126 | brd3a | -0.017 |
| cxcl12a | 0.124 | marcksb | -0.017 |