ADP-ribosylation factor interacting protein 1 (arfaptin 1)
ZFIN 
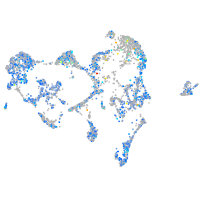
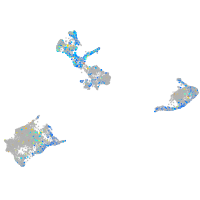
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| qki2 | 0.097 | elavl3 | -0.096 |
| sox2 | 0.093 | myt1b | -0.082 |
| id1 | 0.093 | stxbp1a | -0.080 |
| ntn1b | 0.092 | stx1b | -0.077 |
| fabp7a | 0.091 | elavl4 | -0.077 |
| spon1b | 0.090 | stmn1b | -0.076 |
| wnt4b | 0.090 | gng3 | -0.076 |
| sparc | 0.090 | sncb | -0.074 |
| ctgfa | 0.090 | ptmaa | -0.073 |
| metrnla | 0.090 | snap25a | -0.073 |
| ptn | 0.089 | vamp2 | -0.071 |
| ptprfb | 0.088 | cplx2 | -0.070 |
| si:ch1073-303k11.2 | 0.088 | ptmab | -0.070 |
| anxa13 | 0.087 | syt2a | -0.069 |
| cd82a | 0.087 | onecut1 | -0.069 |
| wls | 0.085 | nsg2 | -0.068 |
| igfbp7 | 0.084 | tmsb | -0.068 |
| hepacama | 0.084 | gng2 | -0.066 |
| gpc5b | 0.083 | jagn1a | -0.066 |
| si:ch211-286b5.5 | 0.082 | rtn1b | -0.066 |
| shha | 0.082 | rtn1a | -0.065 |
| cd63 | 0.082 | tmem59l | -0.065 |
| slit2 | 0.081 | scrt2 | -0.065 |
| rpz5 | 0.080 | stmn2a | -0.064 |
| rfx4 | 0.080 | LOC100537384 | -0.064 |
| npc2 | 0.080 | rbfox1 | -0.064 |
| vamp3 | 0.080 | ywhag2 | -0.064 |
| fxyd1 | 0.079 | gap43 | -0.063 |
| sulf1 | 0.079 | myt1a | -0.062 |
| pgrmc1 | 0.079 | cplx2l | -0.062 |
| atp1b4 | 0.079 | onecut2 | -0.061 |
| slit1b | 0.078 | epb41a | -0.060 |
| ston2 | 0.077 | id4 | -0.060 |
| cyr61l2 | 0.077 | rbfox3a | -0.060 |
| cavin1b | 0.077 | atp6v0cb | -0.060 |