ADP-ribosylation factor 1
ZFIN 
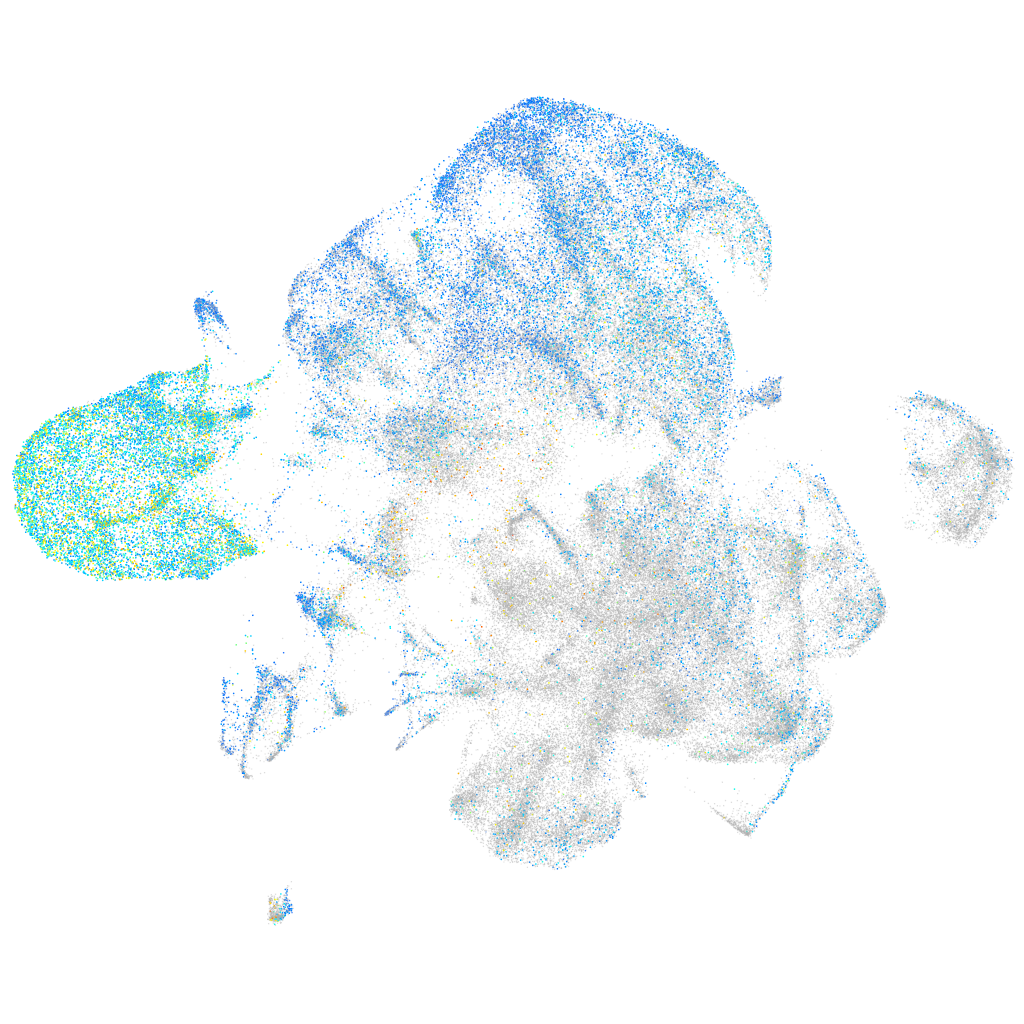
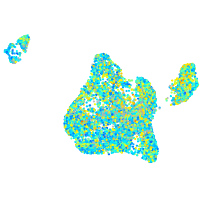
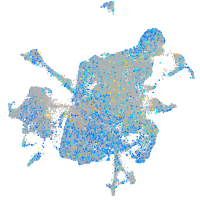
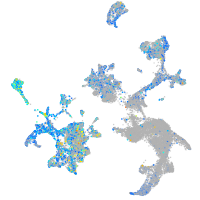
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hspb1 | 0.537 | ptmaa | -0.410 |
| si:ch211-152c2.3 | 0.535 | rpl37 | -0.378 |
| apoeb | 0.499 | rps10 | -0.373 |
| stm | 0.498 | zgc:114188 | -0.356 |
| pou5f3 | 0.460 | h3f3a | -0.348 |
| apoc1 | 0.457 | tuba1c | -0.335 |
| polr3gla | 0.452 | CR383676.1 | -0.310 |
| NC-002333.4 | 0.451 | marcksl1a | -0.304 |
| fbl | 0.447 | hnrnpa0l | -0.300 |
| crabp2b | 0.440 | ppiab | -0.297 |
| npm1a | 0.436 | rtn1a | -0.291 |
| dkc1 | 0.434 | gpm6aa | -0.290 |
| s100a1 | 0.431 | tmsb4x | -0.279 |
| nop58 | 0.430 | stmn1b | -0.278 |
| zmp:0000000624 | 0.421 | elavl3 | -0.270 |
| si:dkey-66i24.9 | 0.421 | fabp3 | -0.256 |
| akap12b | 0.411 | si:ch1073-429i10.3.1 | -0.250 |
| anp32e | 0.404 | gpm6ab | -0.249 |
| apela | 0.402 | ckbb | -0.249 |
| gar1 | 0.401 | calm1a | -0.244 |
| nop2 | 0.401 | rps17 | -0.243 |
| nop56 | 0.400 | zgc:158463 | -0.240 |
| ncl | 0.399 | rnasekb | -0.238 |
| bms1 | 0.398 | gng3 | -0.236 |
| gnl3 | 0.398 | atp6v0cb | -0.236 |
| nr6a1a | 0.397 | nova2 | -0.233 |
| aldob | 0.396 | sncb | -0.232 |
| hnrnpa1b | 0.395 | ppdpfb | -0.230 |
| ddx18 | 0.391 | tmsb | -0.228 |
| lig1 | 0.387 | h3f3c | -0.227 |
| si:ch1073-80i24.3 | 0.386 | mt-nd1 | -0.227 |
| rsl1d1 | 0.385 | gapdhs | -0.226 |
| LOC108190024 | 0.381 | celf2 | -0.226 |
| ebna1bp2 | 0.380 | ccni | -0.226 |
| COX7A2 | 0.377 | COX3 | -0.221 |