ADP-ribosylation factor 1
ZFIN 
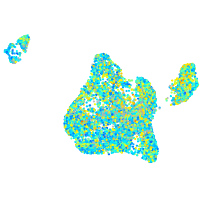
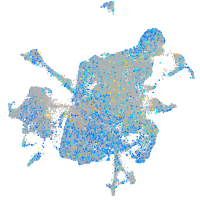
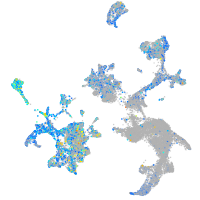
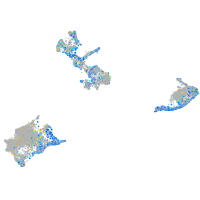
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hspb1 | 0.522 | rpl37 | -0.433 |
| akap12b | 0.520 | rps10 | -0.430 |
| hnrnpa1a | 0.493 | zgc:114188 | -0.405 |
| marcksb | 0.489 | gamt | -0.395 |
| hnrnpa1b | 0.489 | ahcy | -0.385 |
| si:ch211-152c2.3 | 0.485 | gapdh | -0.364 |
| hnrnpub | 0.484 | apoa1b | -0.348 |
| acin1a | 0.480 | bhmt | -0.348 |
| cx43.4 | 0.476 | rps17 | -0.344 |
| hnrnpm | 0.474 | suclg1 | -0.338 |
| NC-002333.4 | 0.468 | eno3 | -0.325 |
| marcksl1b | 0.463 | gatm | -0.325 |
| nucks1a | 0.463 | apoa2 | -0.325 |
| ilf3b | 0.462 | pklr | -0.322 |
| snrnp70 | 0.461 | gpx4a | -0.322 |
| asph | 0.458 | zgc:92744 | -0.322 |
| syncrip | 0.454 | nme2b.1 | -0.317 |
| ppig | 0.452 | apoa4b.1 | -0.313 |
| vox | 0.452 | mat1a | -0.312 |
| rbm4.3 | 0.451 | grhprb | -0.312 |
| sp5l | 0.447 | prdx2 | -0.308 |
| stm | 0.444 | fbp1b | -0.306 |
| smarca4a | 0.444 | afp4 | -0.302 |
| ctnnb1 | 0.444 | agxtb | -0.301 |
| anp32e | 0.442 | apoc2 | -0.301 |
| brd3a | 0.440 | tpi1b | -0.297 |
| top1l | 0.436 | sod1 | -0.295 |
| hmga1a | 0.433 | scp2a | -0.294 |
| smc1al | 0.432 | fabp3 | -0.292 |
| srrm1 | 0.431 | agxta | -0.292 |
| safb | 0.431 | serpina1 | -0.291 |
| sox32 | 0.429 | zgc:123103 | -0.290 |
| anp32a | 0.427 | rbp4 | -0.288 |
| si:ch73-281n10.2 | 0.427 | fabp10a | -0.287 |
| cxcr4a | 0.426 | rbp2b | -0.287 |