"apoptogenic 1, mitochondrial [Source:ZFIN;Acc:ZDB-GENE-090313-66]"
ZFIN 
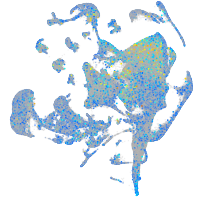
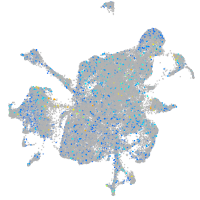
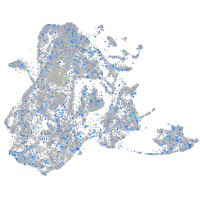
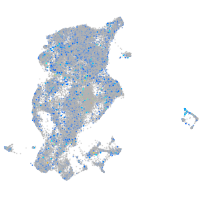
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ccl36.1 | 0.254 | seta | -0.142 |
| cdh17 | 0.203 | ptmab | -0.134 |
| nav2a | 0.193 | hnrnpaba | -0.133 |
| atp1b1a | 0.191 | wu:fb97g03 | -0.132 |
| LOC103909566 | 0.179 | hmgb1b | -0.131 |
| si:ch211-127d4.3 | 0.179 | cirbpa | -0.129 |
| uqcrc2a | 0.177 | si:ch73-281n10.2 | -0.128 |
| atp5f1b | 0.176 | apoeb | -0.128 |
| or125-4 | 0.175 | marcksb | -0.126 |
| si:dkeyp-68b7.12 | 0.174 | cx43.4 | -0.125 |
| vdac2 | 0.174 | si:ch73-1a9.3 | -0.125 |
| arhgap27 | 0.174 | hmga1a | -0.124 |
| slc5a6b | 0.172 | hspb1 | -0.121 |
| atp5if1b | 0.172 | tubb2b | -0.121 |
| atp1a1a.4 | 0.171 | cirbpb | -0.119 |
| atp5meb | 0.168 | setb | -0.119 |
| skap2 | 0.167 | marcksl1b | -0.118 |
| dldh | 0.165 | ncl | -0.118 |
| si:ch1073-325m22.2 | 0.161 | npm1a | -0.118 |
| slc25a5 | 0.160 | hmgn7 | -0.116 |
| glud1b | 0.160 | akap12b | -0.116 |
| zgc:165573 | 0.159 | anp32a | -0.116 |
| bin2a | 0.159 | hmgn2 | -0.114 |
| zgc:56622 | 0.159 | top1l | -0.114 |
| atp5f1d | 0.158 | cdx4 | -0.114 |
| cox6a1 | 0.158 | si:dkey-151g10.6 | -0.112 |
| ndufa5 | 0.158 | syncrip | -0.112 |
| ndufa4l | 0.158 | ilf3b | -0.111 |
| etnk1 | 0.157 | cbx3a | -0.111 |
| si:dkey-16p21.8 | 0.157 | mphosph10 | -0.111 |
| si:dkey-33c14.6 | 0.157 | hnrnpa1a | -0.110 |
| gcdhb | 0.156 | apoc1 | -0.110 |
| chchd10 | 0.155 | h3f3d | -0.109 |
| zgc:193541 | 0.154 | cxcl12a | -0.109 |
| ppifb | 0.151 | chaf1a | -0.107 |