apolipoprotein A-Ib
ZFIN 
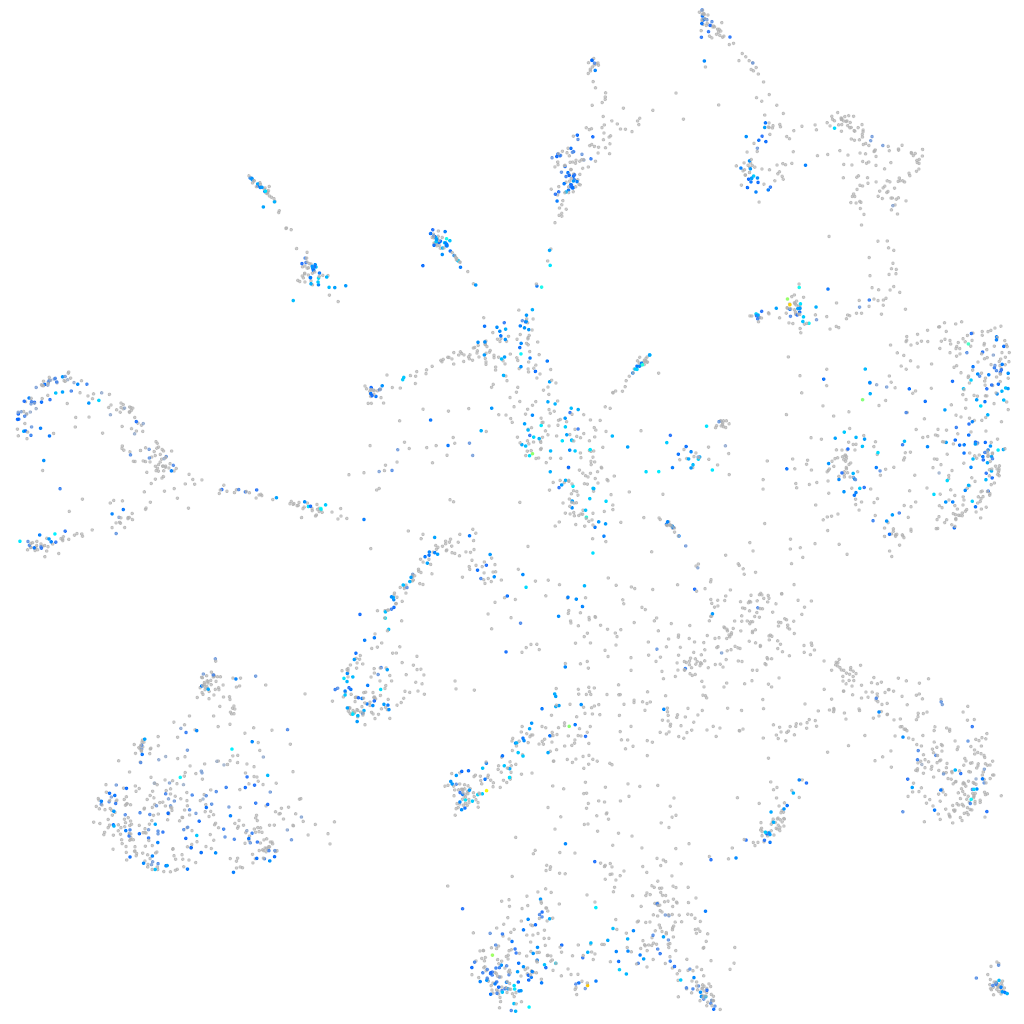
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| apoa2 | 0.438 | rps12 | -0.225 |
| pvalb2 | 0.301 | hmga2 | -0.218 |
| pvalb1 | 0.300 | si:ch211-222l21.1 | -0.212 |
| prss1 | 0.275 | rplp2l | -0.204 |
| prss59.2 | 0.258 | rpl23a | -0.191 |
| ctrb1 | 0.256 | snrpd1 | -0.188 |
| cebpd | 0.247 | ptmab | -0.187 |
| CELA1 (1 of many) | 0.245 | nucks1a | -0.185 |
| fabp10a | 0.243 | hmga1a | -0.182 |
| mylz3 | 0.236 | rpl5b | -0.180 |
| actc1b | 0.235 | hnrnpaba | -0.178 |
| cyt1 | 0.231 | calr3b | -0.177 |
| rbp4 | 0.224 | cirbpa | -0.177 |
| krt17 | 0.221 | si:ch73-46j18.5 | -0.175 |
| si:dkey-33c14.3 | 0.212 | hmgn6 | -0.174 |
| prss59.1 | 0.206 | rps6 | -0.173 |
| rbp2b | 0.202 | tubb2b | -0.172 |
| krt4 | 0.200 | si:ch73-1a9.3 | -0.172 |
| ifitm1 | 0.199 | h2afvb | -0.172 |
| mylpfa | 0.194 | rpl22l1 | -0.171 |
| tpma | 0.186 | syncrip | -0.171 |
| apoc1 | 0.185 | top1l | -0.170 |
| zgc:136461 | 0.184 | snrpg | -0.170 |
| sycn.2 | 0.181 | cfl1 | -0.169 |
| ier2b | 0.175 | uba52 | -0.168 |
| zgc:123103 | 0.172 | ddost | -0.165 |
| ela2l | 0.172 | rpl15 | -0.165 |
| icn | 0.170 | snrpb | -0.164 |
| nme2b.2 | 0.170 | hmgn2 | -0.164 |
| apoa4b.1 | 0.169 | rpl36 | -0.163 |
| gapdh | 0.169 | ppib | -0.163 |
| ctrl | 0.164 | pabpc1a | -0.162 |
| krt91 | 0.163 | seta | -0.161 |
| fabp2 | 0.162 | calr | -0.160 |
| tnnt3b | 0.162 | hnrnpa0b | -0.159 |