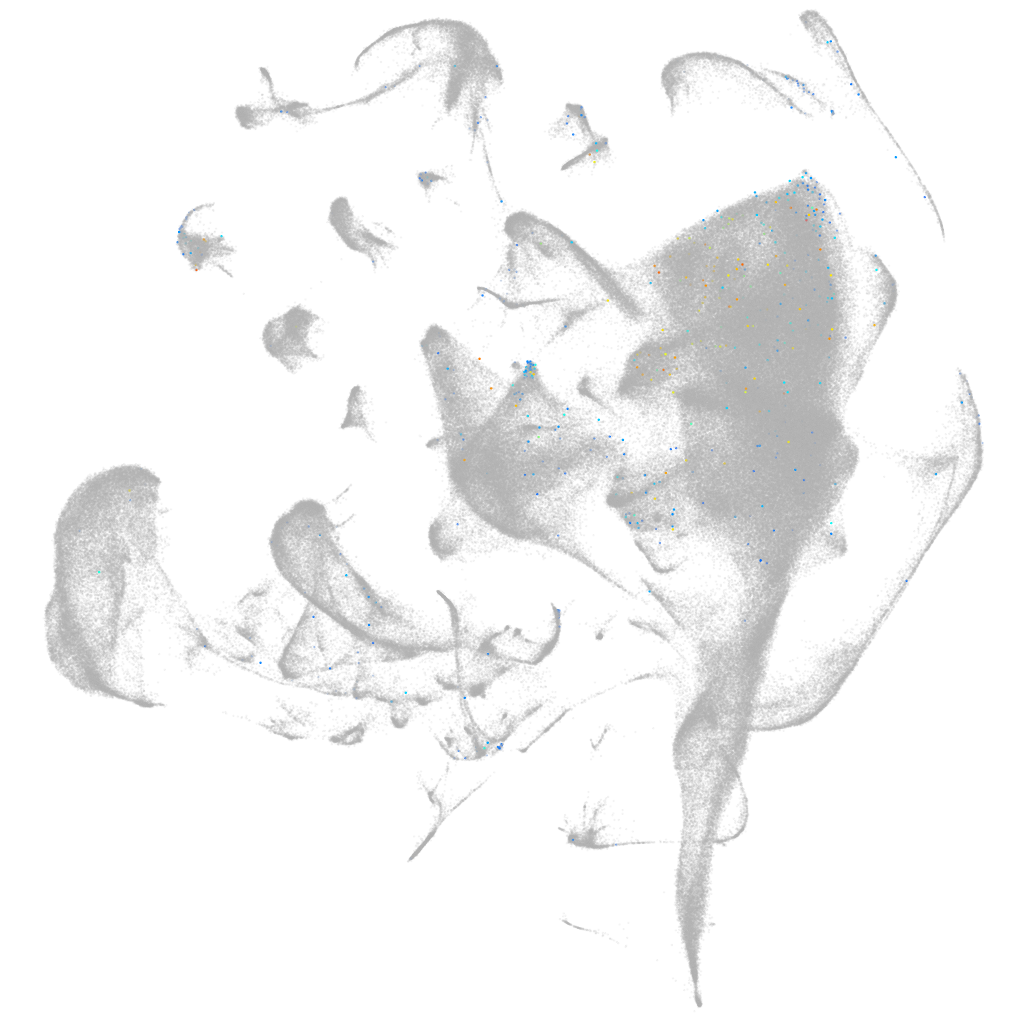
ANTXR cell adhesion molecule 2b
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia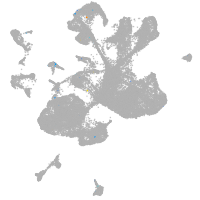 |
eye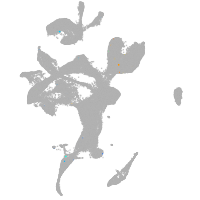 |
taste / olfactory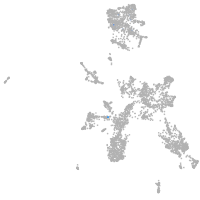 |
otic / lateral line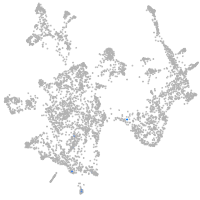 |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| desmb | 0.055 | cx43.4 | -0.011 |
| pdlim3a | 0.049 | ccnb1 | -0.009 |
| csrp1b | 0.049 | chaf1a | -0.009 |
| cnn1b | 0.047 | dkc1 | -0.009 |
| fhl1a | 0.046 | fbl | -0.009 |
| mylkb | 0.046 | mki67 | -0.009 |
| si:ch211-62a1.3 | 0.043 | nop58 | -0.009 |
| acta2 | 0.040 | seta | -0.009 |
| pln | 0.040 | si:ch211-152c2.3 | -0.009 |
| lmod1b | 0.039 | ube2c | -0.009 |
| scn1laa | 0.039 | zic2b | -0.009 |
| si:dkeyp-41f9.4 | 0.039 | aep1 | -0.008 |
| BX323087.1 | 0.038 | aldob | -0.008 |
| myh11a | 0.038 | anp32b | -0.008 |
| smtna | 0.038 | anxa1c | -0.008 |
| tagln | 0.038 | apoc1 | -0.008 |
| si:dkeyp-66d1.7 | 0.037 | banf1 | -0.008 |
| fhl2a | 0.036 | BX927258.1 | -0.008 |
| myocd | 0.035 | ccna2 | -0.008 |
| flnca | 0.033 | cd2bp2 | -0.008 |
| pgm5 | 0.033 | cdca7a | -0.008 |
| tpm1 | 0.032 | cdca7b | -0.008 |
| bves | 0.031 | cyt1 | -0.008 |
| si:ch211-137i24.10 | 0.031 | cyt1l | -0.008 |
| si:dkeyp-51f12.2 | 0.031 | ddx18 | -0.008 |
| cald1b | 0.031 | dla | -0.008 |
| XLOC-041870 | 0.031 | dlgap5 | -0.008 |
| prph | 0.030 | dnmt3bb.2 | -0.008 |
| BX901920.1 | 0.030 | gnl3 | -0.008 |
| myl9a | 0.029 | hnrnpa1b | -0.008 |
| sctr | 0.029 | hnrnpub | -0.008 |
| xgb | 0.029 | kmt5ab | -0.008 |
| pdlim3b | 0.028 | lig1 | -0.008 |
| si:dkey-96g2.1 | 0.028 | lrwd1 | -0.008 |
| sync | 0.028 | lye | -0.008 |