ankyrin repeat and sterile alpha motif domain containing 1B
ZFIN 
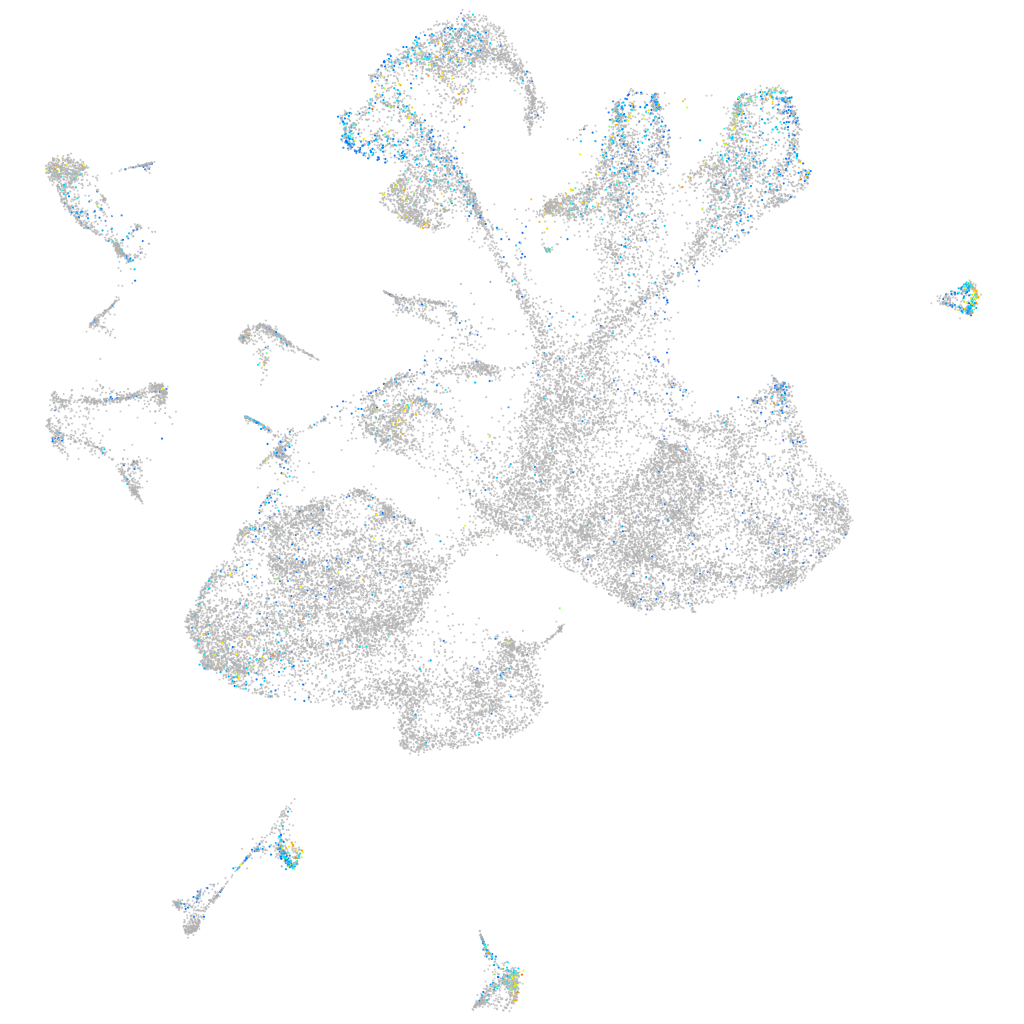
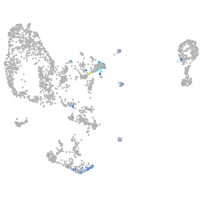
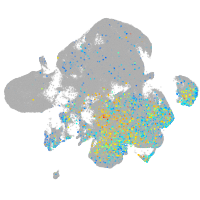
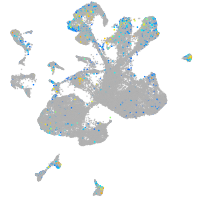
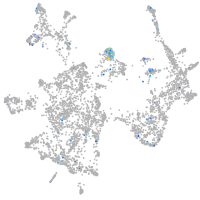
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| pkd2l1 | 0.228 | rplp1 | -0.193 |
| scg2a | 0.219 | si:dkey-151g10.6 | -0.188 |
| scg2b | 0.215 | rps20 | -0.177 |
| cpe | 0.212 | rplp2l | -0.177 |
| pkd1l2a | 0.210 | rpl23 | -0.168 |
| stx1b | 0.208 | rps2 | -0.167 |
| vamp2 | 0.207 | rps7 | -0.167 |
| spock3 | 0.203 | rpl12 | -0.166 |
| snap25a | 0.203 | rps19 | -0.165 |
| sypa | 0.201 | rps15a | -0.163 |
| eno1a | 0.199 | rpl10a | -0.162 |
| stxbp1a | 0.198 | rpl37 | -0.162 |
| calm1b | 0.198 | rps24 | -0.161 |
| ywhag2 | 0.197 | rpl39 | -0.161 |
| zgc:65894 | 0.196 | rps14 | -0.161 |
| si:ch211-113g11.6 | 0.195 | hmgb2a | -0.161 |
| gng3 | 0.193 | rpl11 | -0.160 |
| egr4 | 0.193 | rpl7a | -0.160 |
| sncb | 0.191 | rpl21 | -0.158 |
| si:ch211-214j24.9 | 0.191 | rps12 | -0.158 |
| sncgb | 0.191 | rps5 | -0.157 |
| eno2 | 0.191 | rps9 | -0.157 |
| atp6v0cb | 0.190 | rpl26 | -0.157 |
| sypb | 0.189 | rpl35a | -0.155 |
| sv2a | 0.188 | her15.1 | -0.155 |
| syngr3b | 0.187 | XLOC-003690 | -0.154 |
| gad1b | 0.185 | rpl8 | -0.153 |
| myo3b | 0.185 | hmgb2b | -0.153 |
| vgf | 0.185 | rpsa | -0.153 |
| pcsk1nl | 0.184 | rps3a | -0.151 |
| stmn2a | 0.184 | rpl27 | -0.150 |
| eps8l2 | 0.182 | rpl7 | -0.150 |
| atp1a3a | 0.181 | hmga1a | -0.150 |
| gad2 | 0.179 | rps10 | -0.150 |
| elavl4 | 0.178 | rps27.1 | -0.149 |