ankyrin repeat domain 52a
ZFIN 
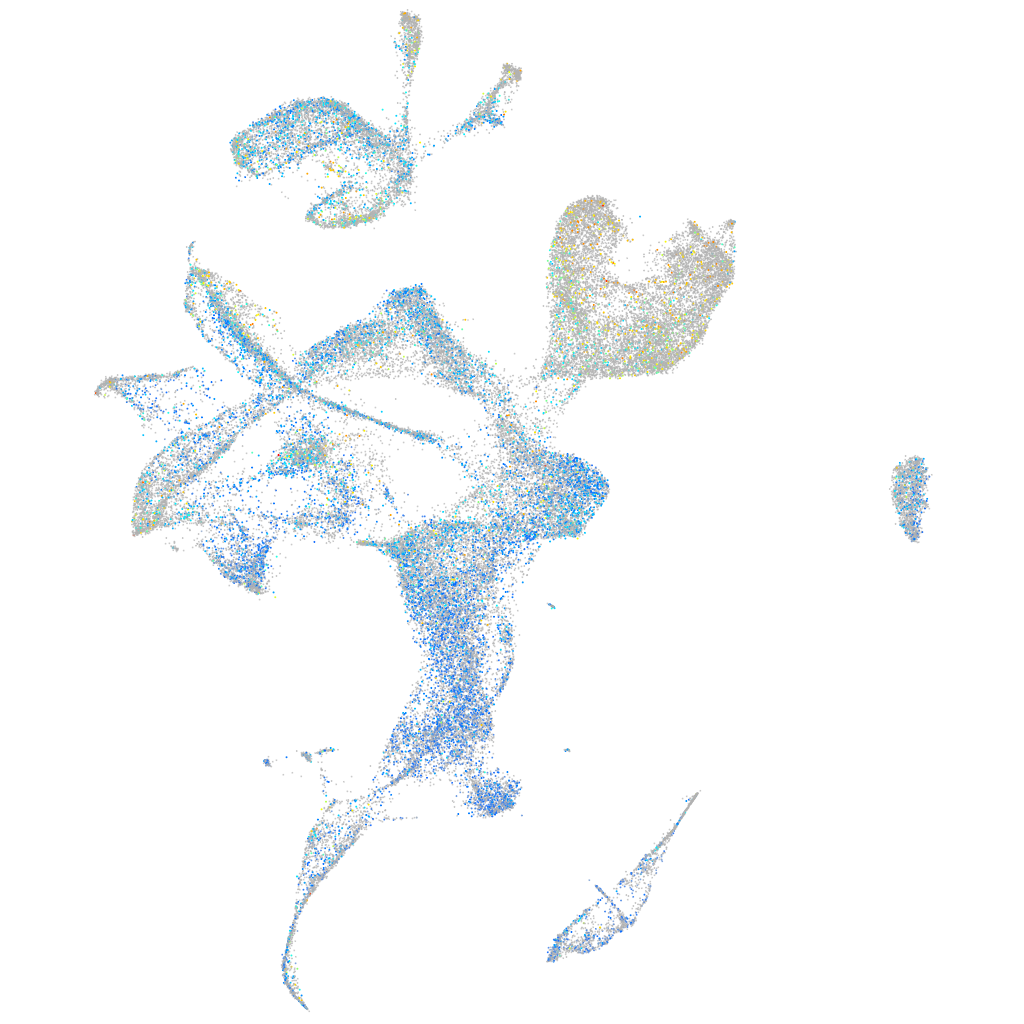
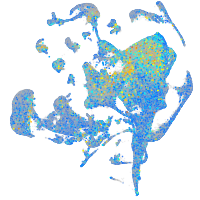
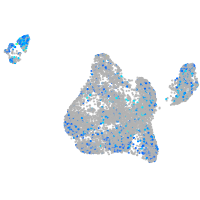
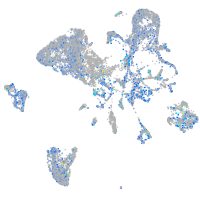
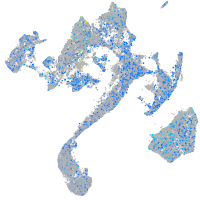
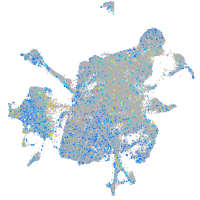
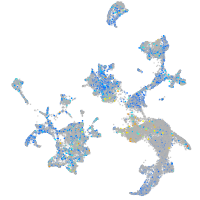
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| celf2 | 0.088 | syt5b | -0.043 |
| tuba1a | 0.078 | nrn1lb | -0.040 |
| tubb5 | 0.069 | si:dkey-16p21.8 | -0.038 |
| tmpob | 0.067 | gnb3a | -0.036 |
| elavl3 | 0.067 | actc1b | -0.035 |
| gnb1b | 0.067 | pvalb1 | -0.035 |
| tuba1c | 0.066 | vamp1 | -0.035 |
| hnrnpa0a | 0.065 | si:ch73-256g18.2 | -0.033 |
| ctbp1 | 0.064 | cabp2a | -0.033 |
| actb1 | 0.064 | snx8b | -0.033 |
| eef2b | 0.063 | pvalb2 | -0.032 |
| atrx | 0.063 | stx3a | -0.032 |
| tubb2b | 0.062 | rs1a | -0.031 |
| elavl4 | 0.062 | cplx4a | -0.031 |
| pfn2 | 0.062 | lin7a | -0.031 |
| marcksb | 0.061 | dusp2 | -0.030 |
| sinhcafl | 0.061 | si:ch1073-83n3.2 | -0.029 |
| hnrnpaba | 0.060 | npdc1a | -0.029 |
| khdrbs1a | 0.060 | apoa2 | -0.029 |
| rpl3 | 0.060 | calb2b | -0.028 |
| cenpf | 0.059 | gem | -0.028 |
| si:ch211-137a8.4 | 0.059 | prkcaa | -0.028 |
| igf2bp1 | 0.059 | prss1 | -0.027 |
| mt-co1 | 0.058 | tnnt3b | -0.027 |
| syncrip | 0.058 | mylpfa | -0.027 |
| seta | 0.057 | samsn1a | -0.027 |
| aspm | 0.057 | si:ch211-56a11.2 | -0.027 |
| ckap5 | 0.057 | si:ch211-152c2.3 | -0.026 |
| FO082781.1 | 0.057 | apoa1b | -0.026 |
| eif4a1a | 0.057 | gabrr1 | -0.026 |
| gatad2b | 0.056 | CABZ01038709.1 | -0.026 |
| mab21l1 | 0.056 | gnao1b | -0.026 |
| si:ch211-288g17.3 | 0.056 | histh1l | -0.026 |
| cfl1 | 0.056 | rgs16 | -0.026 |
| gnb1a | 0.056 | gng13b | -0.026 |