adhesion molecule with Ig-like domain 3
ZFIN 
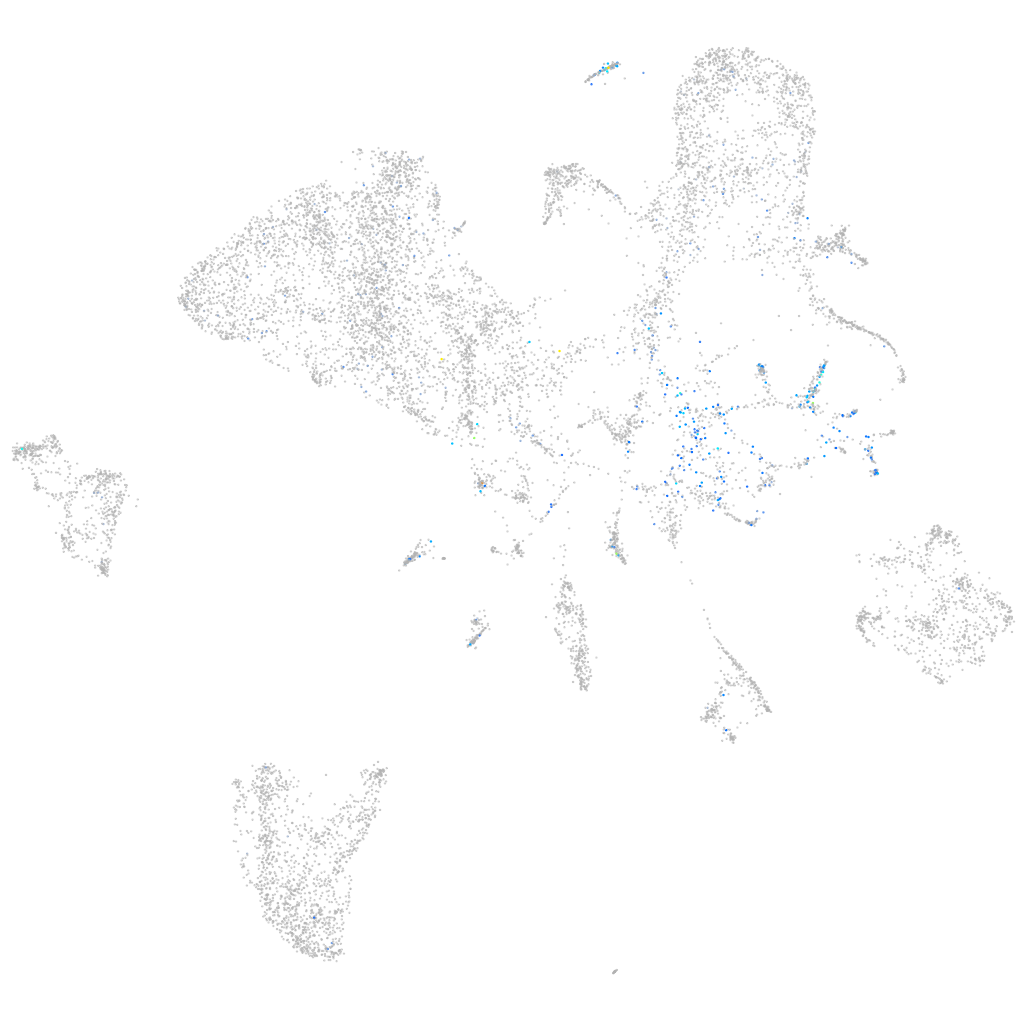
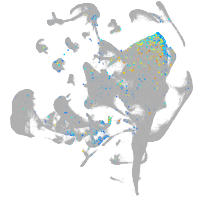
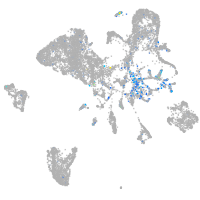
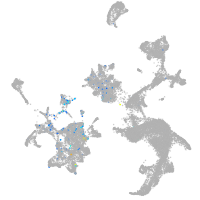
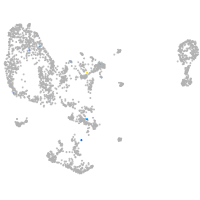
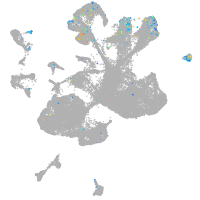
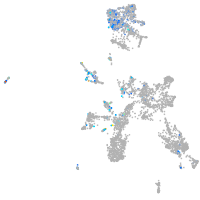
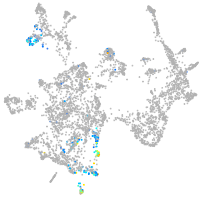
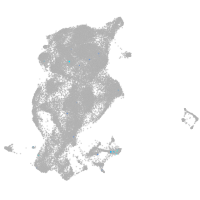
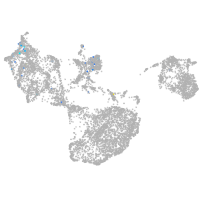
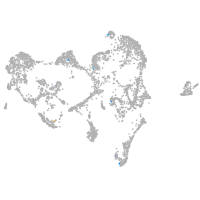
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| scg3 | 0.238 | gapdh | -0.123 |
| egr4 | 0.209 | ahcy | -0.115 |
| neurod1 | 0.205 | gatm | -0.111 |
| vat1 | 0.205 | gamt | -0.109 |
| syt1a | 0.203 | bhmt | -0.101 |
| mir7a-1 | 0.201 | mat1a | -0.099 |
| pcsk1nl | 0.200 | apoa4b.1 | -0.093 |
| rprmb | 0.197 | fbp1b | -0.092 |
| pcsk2 | 0.193 | agxtb | -0.088 |
| dicp1.5-6 | 0.192 | cx32.3 | -0.087 |
| arxa | 0.192 | dap | -0.086 |
| zgc:101731 | 0.190 | gnmt | -0.086 |
| scgn | 0.189 | nupr1b | -0.086 |
| insm1b | 0.186 | afp4 | -0.085 |
| rem2 | 0.185 | apoc2 | -0.085 |
| gcga | 0.183 | mgst1.2 | -0.084 |
| si:dkey-153k10.9 | 0.179 | glud1b | -0.083 |
| fev | 0.179 | apoa1b | -0.082 |
| anxa4 | 0.179 | apoc1 | -0.082 |
| oprd1b | 0.178 | ckba | -0.082 |
| c2cd4a | 0.178 | apobb.1 | -0.081 |
| gper1 | 0.175 | scp2a | -0.080 |
| ccka | 0.171 | pnp4b | -0.080 |
| insl5a | 0.168 | suclg2 | -0.078 |
| rasd4 | 0.168 | gstt1a | -0.078 |
| pax6b | 0.166 | tdo2a | -0.077 |
| nkx2.2a | 0.164 | aldh7a1 | -0.077 |
| rims2a | 0.163 | hdlbpa | -0.076 |
| mir375-2 | 0.161 | aldob | -0.076 |
| elovl1a | 0.160 | etnppl | -0.075 |
| gfra1a | 0.160 | cdo1 | -0.075 |
| or115-9 | 0.160 | ces2 | -0.075 |
| nmbb | 0.160 | fetub | -0.075 |
| pcsk1 | 0.160 | cx28.9 | -0.074 |
| dll4 | 0.157 | apoa2 | -0.074 |