alsin Rho guanine nucleotide exchange factor ALS2 b
ZFIN 
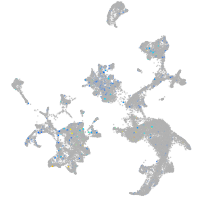
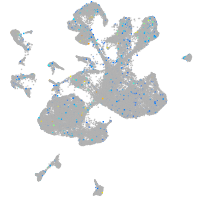
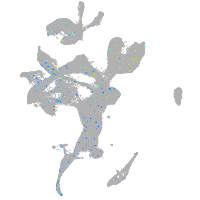
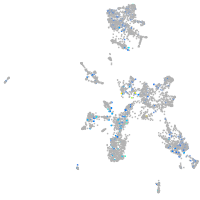
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| intu | 0.673 | gtf2f1 | -0.287 |
| sept8b | 0.661 | ncl | -0.277 |
| fryb | 0.654 | akap12b | -0.264 |
| cers6 | 0.634 | ddx18 | -0.263 |
| kctd6a | 0.613 | hnrnpa1a | -0.262 |
| zswim2 | 0.587 | NC-002333.4 | -0.260 |
| kcnh1b | 0.587 | dlgap5 | -0.243 |
| mapta | 0.570 | srrt | -0.241 |
| si:ch73-22o18.1 | 0.555 | acin1b | -0.240 |
| zgc:77439 | 0.547 | pttg1 | -0.237 |
| FP102018.1 | 0.541 | zgc:110425 | -0.234 |
| trpm4a | 0.540 | thoc7 | -0.229 |
| ano10a | 0.532 | ctcf | -0.229 |
| deptor | 0.531 | nop58 | -0.227 |
| ccdc114 | 0.529 | brd3a | -0.224 |
| ifit12 | 0.526 | si:ch73-281n10.2 | -0.223 |
| slc5a1 | 0.524 | hnrnpub | -0.223 |
| ano9a | 0.511 | sec62 | -0.220 |
| CU633762.1 | 0.506 | gra | -0.218 |
| slc37a4b | 0.503 | hnrnpa1b | -0.213 |
| celsr1b | 0.494 | ebna1bp2 | -0.213 |
| zcchc14 | 0.488 | tpx2 | -0.212 |
| abo | 0.487 | apoeb | -0.211 |
| atg16l2 | 0.487 | fbl | -0.206 |
| BX927130.4 | 0.487 | ppig | -0.205 |
| c1ql3a | 0.487 | knop1 | -0.204 |
| CABZ01066320.2 | 0.487 | marcksb | -0.204 |
| calb1 | 0.487 | snrpb2 | -0.204 |
| CR354610.1 | 0.487 | metap2b | -0.203 |
| CR385024.1 | 0.487 | gar1 | -0.203 |
| cx23 | 0.487 | ccnb1 | -0.201 |
| ftr57 | 0.487 | tbx16 | -0.200 |
| gcdhb | 0.487 | zgc:112356 | -0.200 |
| GRIK2 | 0.487 | marcksl1b | -0.199 |
| itga3a | 0.487 | puf60b | -0.199 |