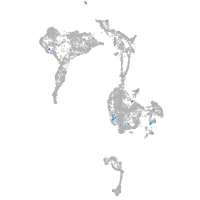
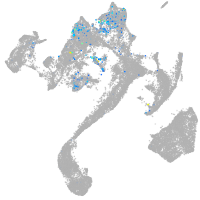
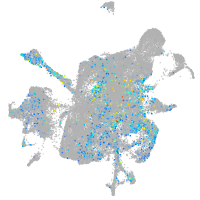
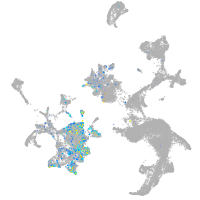
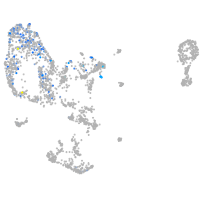
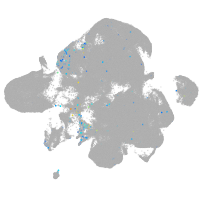
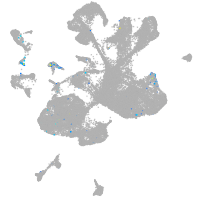
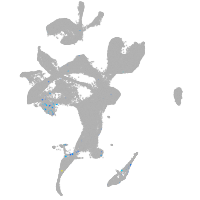
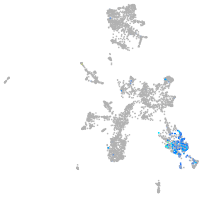
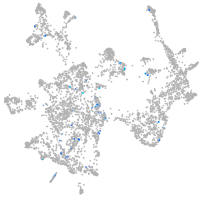
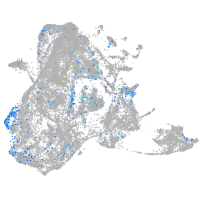
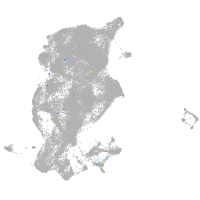
adrenomedullin a
ZFIN 
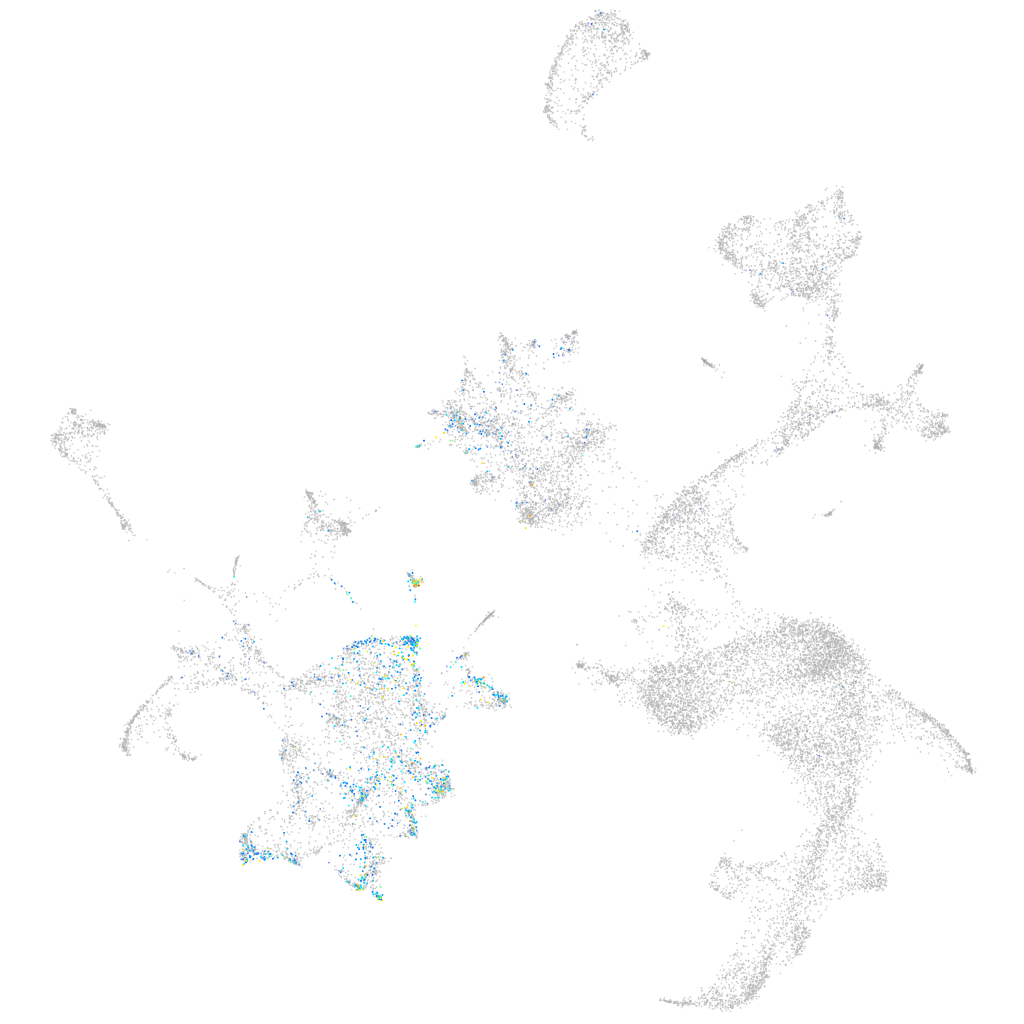
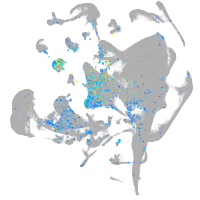
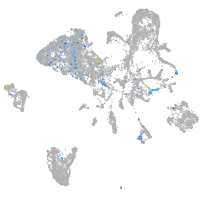
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| igfbp7 | 0.355 | prdx2 | -0.180 |
| cpn1 | 0.351 | hbae1.1 | -0.179 |
| si:ch211-145b13.6 | 0.331 | hbae1.3 | -0.170 |
| plvapb | 0.313 | hbbe2 | -0.165 |
| cldn5b | 0.307 | hbbe1.3 | -0.162 |
| lyve1a | 0.304 | cahz | -0.161 |
| cdh5 | 0.301 | hbae3 | -0.161 |
| tie1 | 0.300 | fth1a | -0.159 |
| krt18a.1 | 0.298 | hbbe1.1 | -0.158 |
| krt8 | 0.292 | blvrb | -0.154 |
| rgcc | 0.290 | hemgn | -0.153 |
| col4a1 | 0.287 | hbbe1.2 | -0.148 |
| tspan4b | 0.285 | alas2 | -0.145 |
| fam174b | 0.283 | slc4a1a | -0.138 |
| pecam1 | 0.279 | nt5c2l1 | -0.134 |
| vwa1 | 0.273 | tspo | -0.133 |
| clec14a | 0.271 | epb41b | -0.131 |
| si:ch211-248e11.2 | 0.270 | zgc:163057 | -0.128 |
| kdr | 0.269 | si:ch211-250g4.3 | -0.128 |
| ednraa | 0.268 | gpx1a | -0.127 |
| myct1a | 0.267 | creg1 | -0.123 |
| ramp2 | 0.265 | si:ch211-207c6.2 | -0.119 |
| plk2a | 0.263 | nmt1b | -0.117 |
| kdrl | 0.262 | plac8l1 | -0.114 |
| plecb | 0.261 | tmod4 | -0.110 |
| tgfbr2b | 0.259 | vdac2 | -0.107 |
| vat1 | 0.258 | znfl2a | -0.106 |
| dll4 | 0.257 | sptb | -0.103 |
| igfbp1a | 0.256 | zgc:56095 | -0.103 |
| si:ch211-33e4.2 | 0.255 | urod | -0.102 |
| sox18 | 0.252 | hbae5 | -0.101 |
| hlx1 | 0.251 | tfr1a | -0.100 |
| scarb2a | 0.251 | hdr | -0.100 |
| notchl | 0.250 | hbbe3 | -0.100 |
| anxa13 | 0.249 | rfesd | -0.100 |