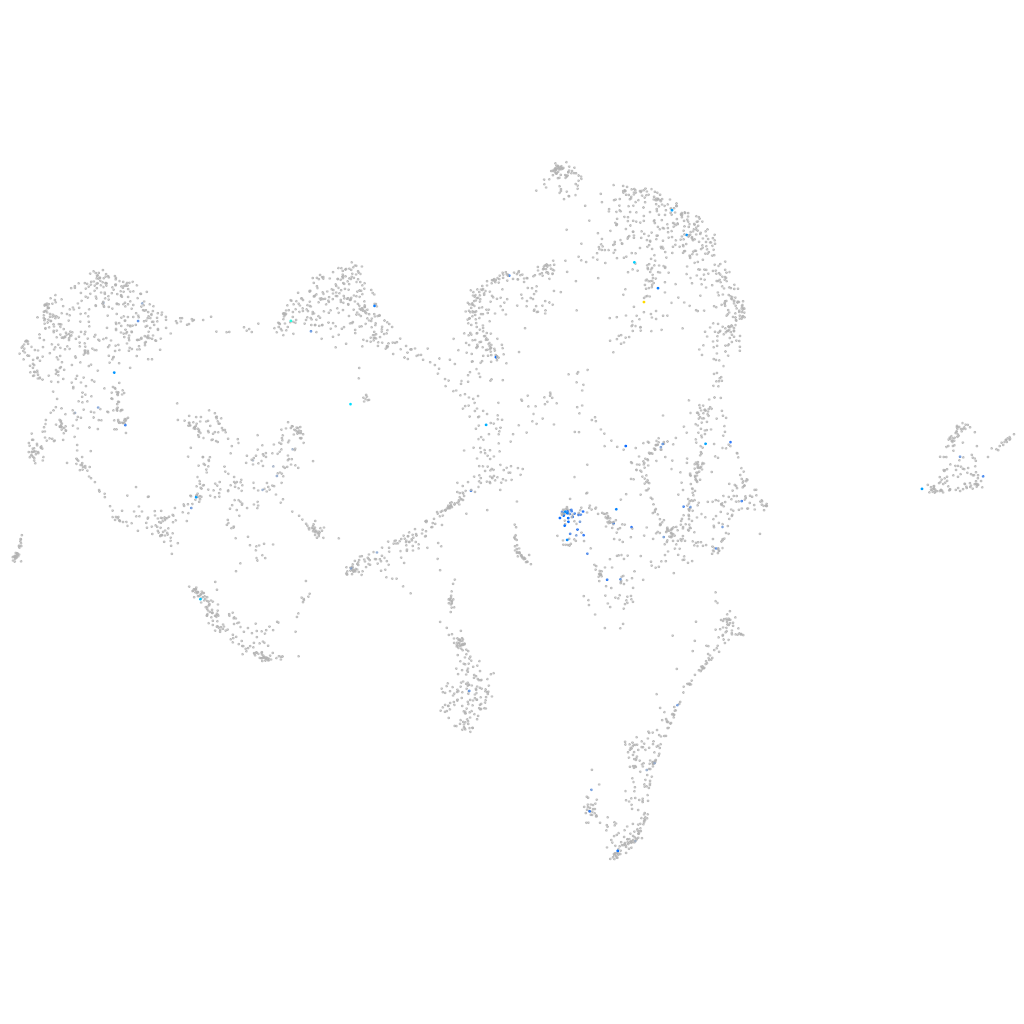
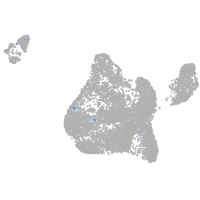
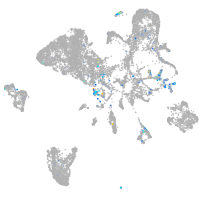
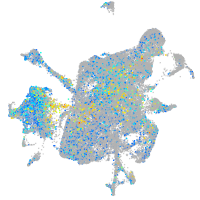
adhesion G protein-coupled receptor L3.1
ZFIN 
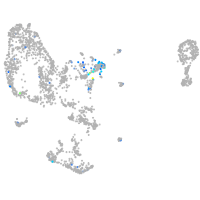
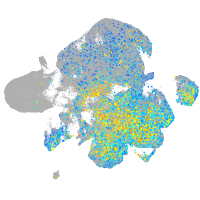
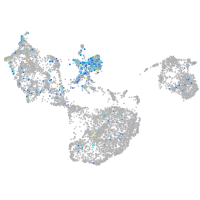
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| lingo1a | 0.336 | pkp3a | -0.076 |
| rhbdl1 | 0.335 | krt4 | -0.067 |
| gja5b | 0.317 | prr13 | -0.058 |
| BX088596.2 | 0.284 | asah1b | -0.057 |
| LOC108182922 | 0.266 | rnaseka | -0.055 |
| LOC110439439 | 0.266 | uqcrc2b | -0.054 |
| sdhda | 0.266 | tm9sf5 | -0.053 |
| sncb | 0.265 | anxa11b | -0.053 |
| gig2i | 0.262 | cst14a.2 | -0.053 |
| tuba1a | 0.261 | zgc:110333 | -0.053 |
| trim35-13 | 0.254 | pkp1b | -0.053 |
| grin1b | 0.238 | CU693379.1 | -0.053 |
| atp6v0cb | 0.236 | ezrb | -0.052 |
| rab42a | 0.231 | prkab1b | -0.052 |
| gng3 | 0.229 | sgk2a | -0.051 |
| stxbp1a | 0.228 | mpc1 | -0.051 |
| stmn1b | 0.228 | fuca1.1 | -0.051 |
| ywhag2 | 0.226 | cldnh | -0.051 |
| tuba1c | 0.223 | pdlim2 | -0.050 |
| ndrg4 | 0.222 | f11r.1 | -0.050 |
| pecam1 | 0.221 | svopl | -0.050 |
| CR376838.1 | 0.218 | b3gnt2l | -0.050 |
| caly | 0.217 | mid1ip1a | -0.049 |
| gja5a | 0.216 | enosf1 | -0.049 |
| FO704648.2 | 0.216 | tmem126a | -0.049 |
| zgc:194906 | 0.215 | pls3 | -0.049 |
| flt1 | 0.214 | hif1al | -0.049 |
| zgc:153426 | 0.213 | waslb | -0.049 |
| sez6b | 0.212 | reep3b | -0.049 |
| onecut3a | 0.211 | dspa | -0.048 |
| ecscr | 0.211 | padi2 | -0.048 |
| rtn1b | 0.210 | ifi30 | -0.048 |
| aff2 | 0.208 | nenf | -0.048 |
| elavl3 | 0.206 | si:dkey-16p21.8 | -0.048 |
| lrrfip1b | 0.206 | hdlbpa | -0.048 |