adhesion G protein-coupled receptor A1a
ZFIN 
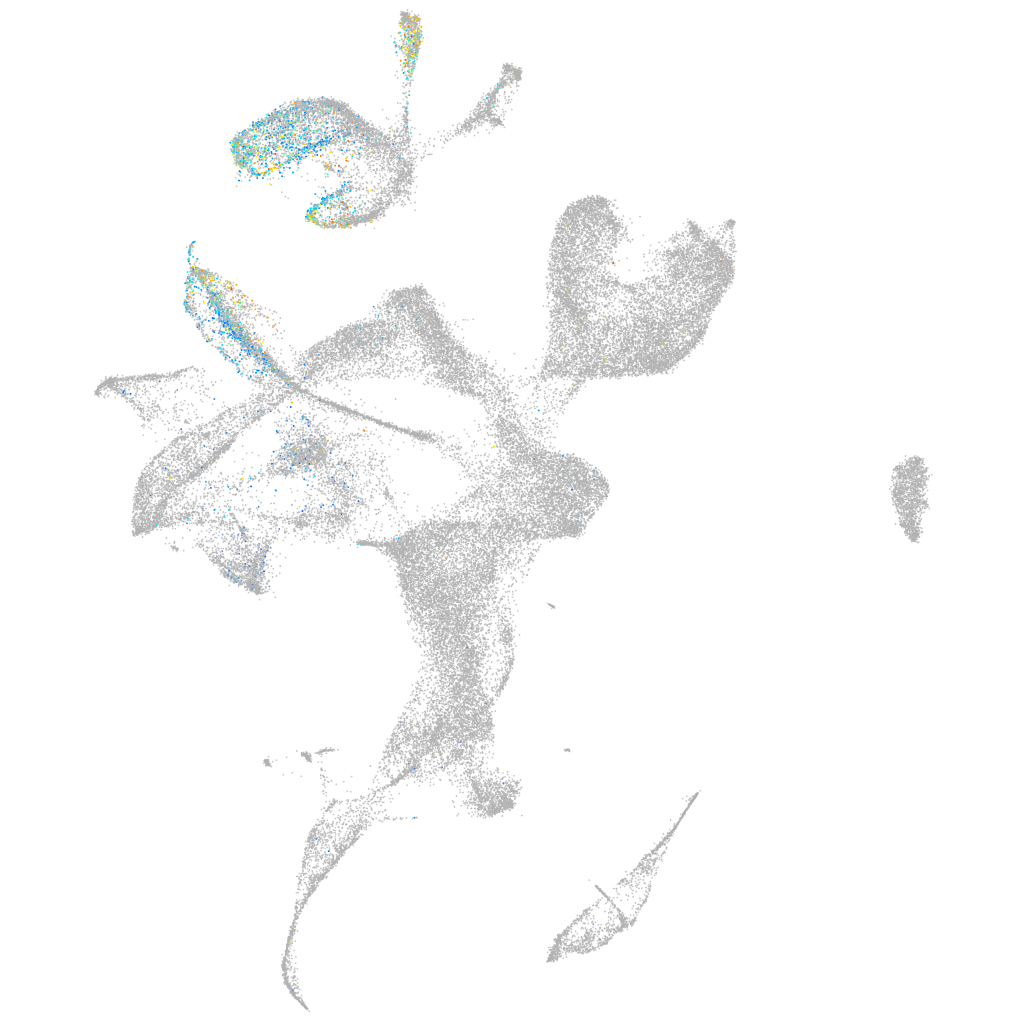
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| stxbp1a | 0.311 | hmgb2a | -0.206 |
| syt1a | 0.306 | hmga1a | -0.118 |
| rbfox1 | 0.300 | mdka | -0.109 |
| sv2a | 0.295 | hmgb2b | -0.105 |
| slc32a1 | 0.293 | stmn1a | -0.105 |
| stx1b | 0.293 | ahcy | -0.105 |
| eno2 | 0.288 | pcna | -0.102 |
| ywhag2 | 0.287 | msi1 | -0.101 |
| gabrb4 | 0.272 | msna | -0.097 |
| grin1a | 0.268 | lbr | -0.095 |
| si:ch73-119p20.1 | 0.266 | dut | -0.095 |
| syt2a | 0.264 | nutf2l | -0.095 |
| sncb | 0.258 | rrm1 | -0.095 |
| zgc:92912 | 0.255 | rps20 | -0.095 |
| tkta | 0.254 | fabp7a | -0.092 |
| gria2a | 0.251 | mki67 | -0.091 |
| st8sia5 | 0.249 | ccnd1 | -0.090 |
| rtn1b | 0.247 | rpl12 | -0.090 |
| si:ch211-129p13.1 | 0.247 | mcm7 | -0.090 |
| nrxn1a | 0.244 | si:dkey-151g10.6 | -0.090 |
| elavl3 | 0.243 | tuba8l | -0.089 |
| syt3 | 0.240 | chaf1a | -0.088 |
| zgc:153426 | 0.237 | rplp2l | -0.088 |
| cabp1b | 0.235 | tuba8l4 | -0.088 |
| rims2a | 0.233 | rpl11 | -0.087 |
| snap25a | 0.231 | COX7A2 (1 of many) | -0.087 |
| mir181b-3 | 0.230 | neurod4 | -0.087 |
| gng3 | 0.224 | her15.1 | -0.087 |
| sprn | 0.224 | rpl39 | -0.087 |
| gnao1a | 0.222 | selenoh | -0.086 |
| plppr4b | 0.221 | eef1da | -0.086 |
| cplx2 | 0.221 | ccna2 | -0.085 |
| zgc:65894 | 0.215 | cx43.4 | -0.084 |
| vsnl1b | 0.215 | tspan7 | -0.084 |
| slc6a1a | 0.215 | rps19 | -0.084 |