ADAM metallopeptidase domain 19a
ZFIN 
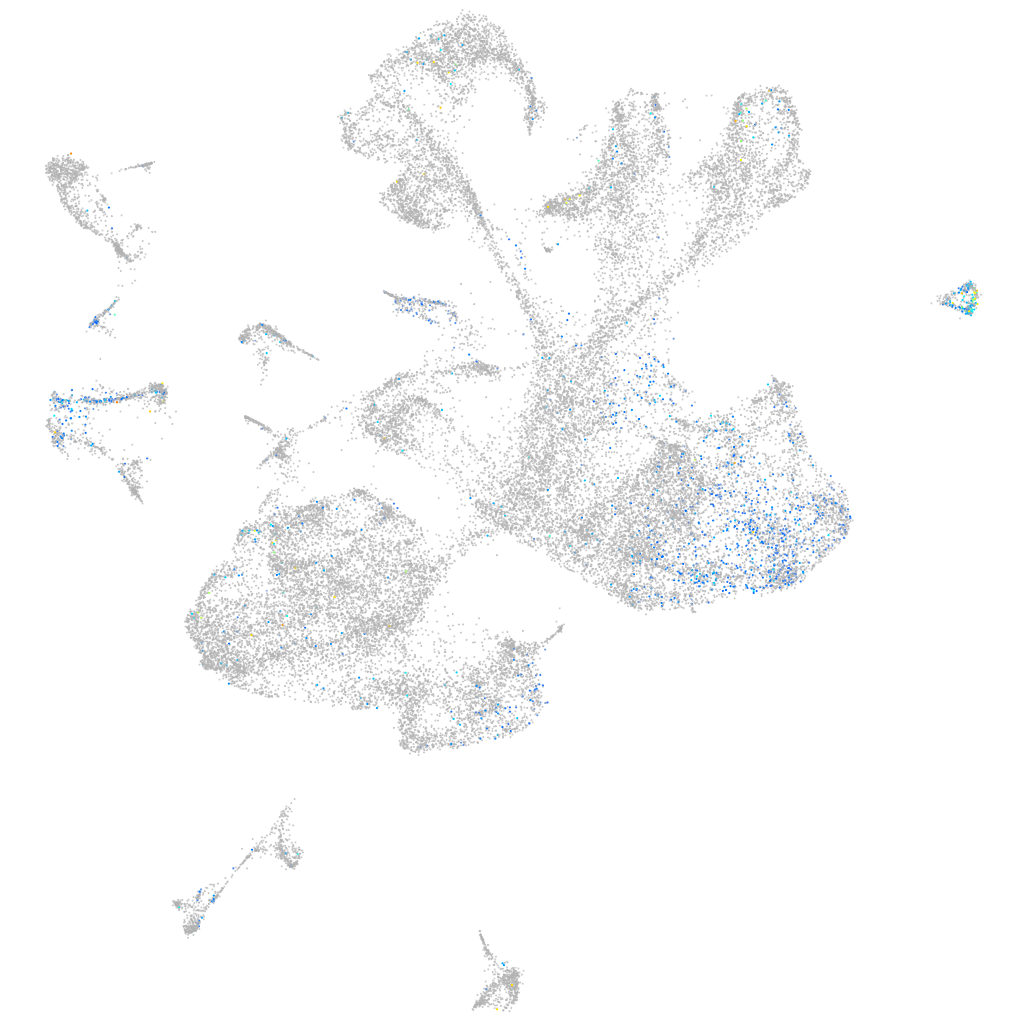
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| pkd1l2a | 0.210 | CU467822.1 | -0.084 |
| pkd2l1 | 0.206 | tmsb4x | -0.078 |
| sst1.1 | 0.179 | tmsb | -0.077 |
| scg2a | 0.176 | CU634008.1 | -0.077 |
| myo3b | 0.171 | zgc:165461 | -0.071 |
| bicdl2l | 0.163 | ckbb | -0.069 |
| oc90 | 0.161 | fabp7a | -0.068 |
| nppc | 0.160 | pvalb1 | -0.066 |
| tac3a | 0.157 | si:ch211-57n23.4 | -0.066 |
| c2cd4a | 0.152 | pvalb2 | -0.066 |
| esm1 | 0.150 | actc1b | -0.063 |
| tal2 | 0.147 | gpm6aa | -0.060 |
| cdx4 | 0.143 | marcksl1a | -0.058 |
| eps8l2 | 0.142 | CR848047.1 | -0.058 |
| ldlrad2 | 0.142 | gpm6bb | -0.057 |
| stac | 0.139 | hmgb3a | -0.057 |
| apela | 0.137 | slc1a2b | -0.055 |
| lrfn4b | 0.135 | fabp3 | -0.055 |
| gad1b | 0.130 | LOC101882472 | -0.054 |
| rhpn2 | 0.129 | COX3 | -0.053 |
| gad2 | 0.128 | hbbe1.3 | -0.053 |
| baiap2l1b | 0.123 | si:ch73-30b17.2 | -0.052 |
| scinlb | 0.121 | BX664610.1 | -0.052 |
| CU856539.2 | 0.120 | si:ch211-251b21.1 | -0.051 |
| si:dkey-30j16.3 | 0.117 | mdkb | -0.051 |
| RASGRF1 | 0.115 | pou3f1 | -0.051 |
| npr3 | 0.115 | pou3f3b | -0.051 |
| baiap2a | 0.114 | myt1b | -0.051 |
| hspb1 | 0.114 | npas1 | -0.050 |
| kcnf1a | 0.109 | mylpfa | -0.050 |
| XLOC-001964 | 0.109 | si:ch73-21g5.7 | -0.049 |
| cgnl1 | 0.108 | FO082781.1 | -0.048 |
| crygn1 | 0.108 | nr2f1a | -0.048 |
| grm2a | 0.106 | atp1a1b | -0.048 |
| map2k1 | 0.106 | dpysl2b | -0.048 |