"actinin, alpha 1"
ZFIN 
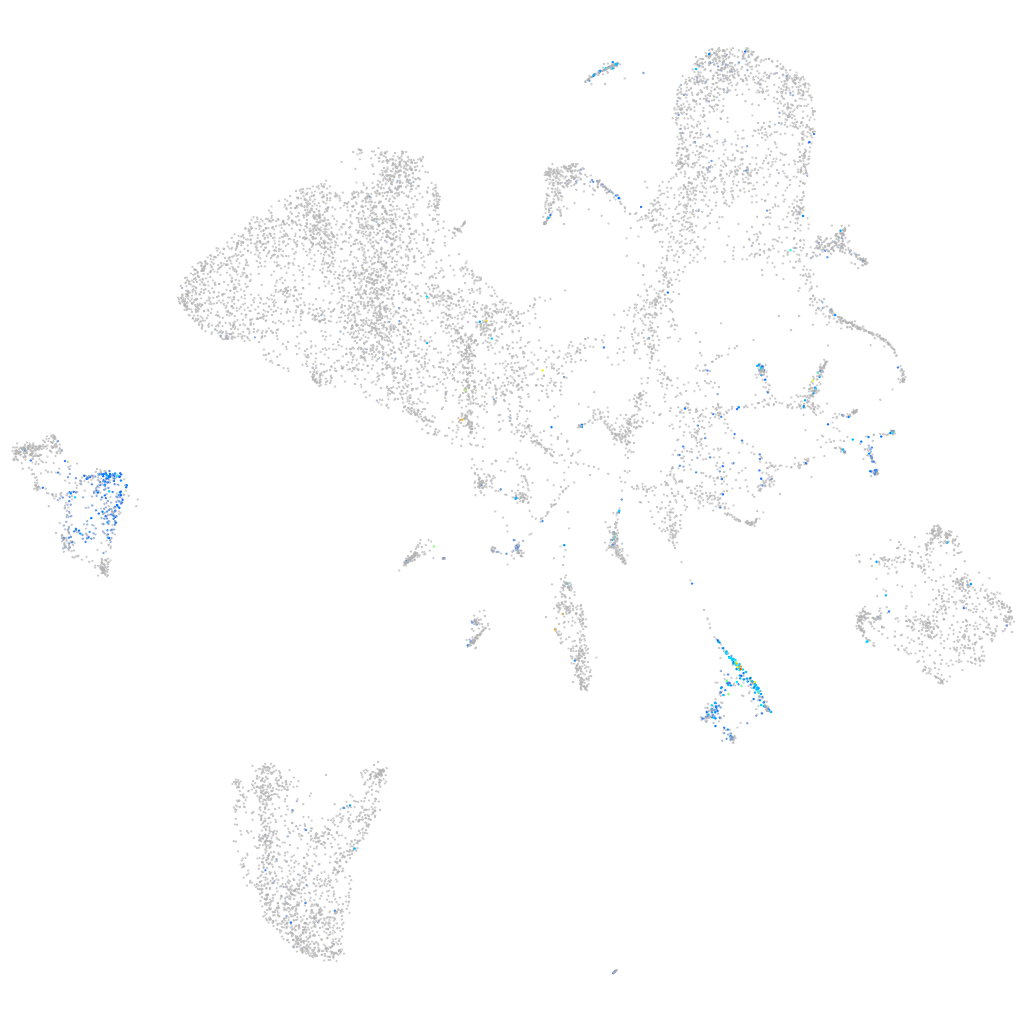
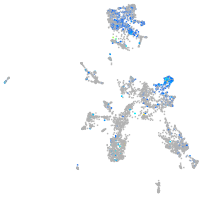
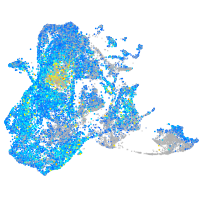
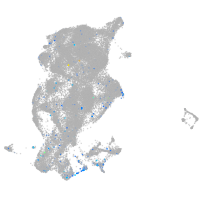
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| pmp22b | 0.423 | gapdh | -0.193 |
| col4a5 | 0.421 | gamt | -0.175 |
| sim1b | 0.418 | gatm | -0.168 |
| abca12 | 0.403 | bhmt | -0.162 |
| lama5 | 0.401 | fbp1b | -0.160 |
| col4a6 | 0.399 | hspe1 | -0.154 |
| vwa1 | 0.388 | apoa4b.1 | -0.153 |
| spock3 | 0.386 | mat1a | -0.153 |
| b3gnt5a | 0.373 | apoc2 | -0.149 |
| sftpba | 0.355 | ahcy | -0.148 |
| sparc | 0.354 | aqp12 | -0.147 |
| CABZ01055365.1 | 0.353 | gpx4a | -0.147 |
| f3a | 0.349 | agxta | -0.146 |
| ahnak | 0.349 | gstt1a | -0.145 |
| cabp2b | 0.346 | scp2a | -0.144 |
| frem2b | 0.342 | apoa1b | -0.144 |
| spaca4l | 0.342 | afp4 | -0.140 |
| anxa3b | 0.332 | apoc1 | -0.139 |
| bcam | 0.332 | agxtb | -0.139 |
| si:ch211-264f5.6 | 0.324 | aldob | -0.136 |
| krt94 | 0.322 | gcshb | -0.135 |
| anxa1a | 0.318 | grhprb | -0.134 |
| spns2 | 0.317 | tdo2a | -0.133 |
| tgfb3 | 0.314 | dap | -0.133 |
| cav1 | 0.313 | pnp4b | -0.132 |
| wnt11r | 0.313 | sult2st2 | -0.131 |
| capn2a | 0.312 | g6pca.2 | -0.129 |
| marcksl1a | 0.305 | msrb2 | -0.129 |
| RASA2 | 0.303 | nipsnap3a | -0.128 |
| gapdhs | 0.303 | upb1 | -0.128 |
| col18a1a | 0.302 | slc27a2a | -0.128 |
| sgms1 | 0.301 | suclg1 | -0.128 |
| ceacam1 | 0.300 | apom | -0.127 |
| cavin2a | 0.295 | ftcd | -0.127 |
| myofl | 0.292 | serpina1 | -0.127 |