acyl-CoA synthetase short chain family member 1
ZFIN 
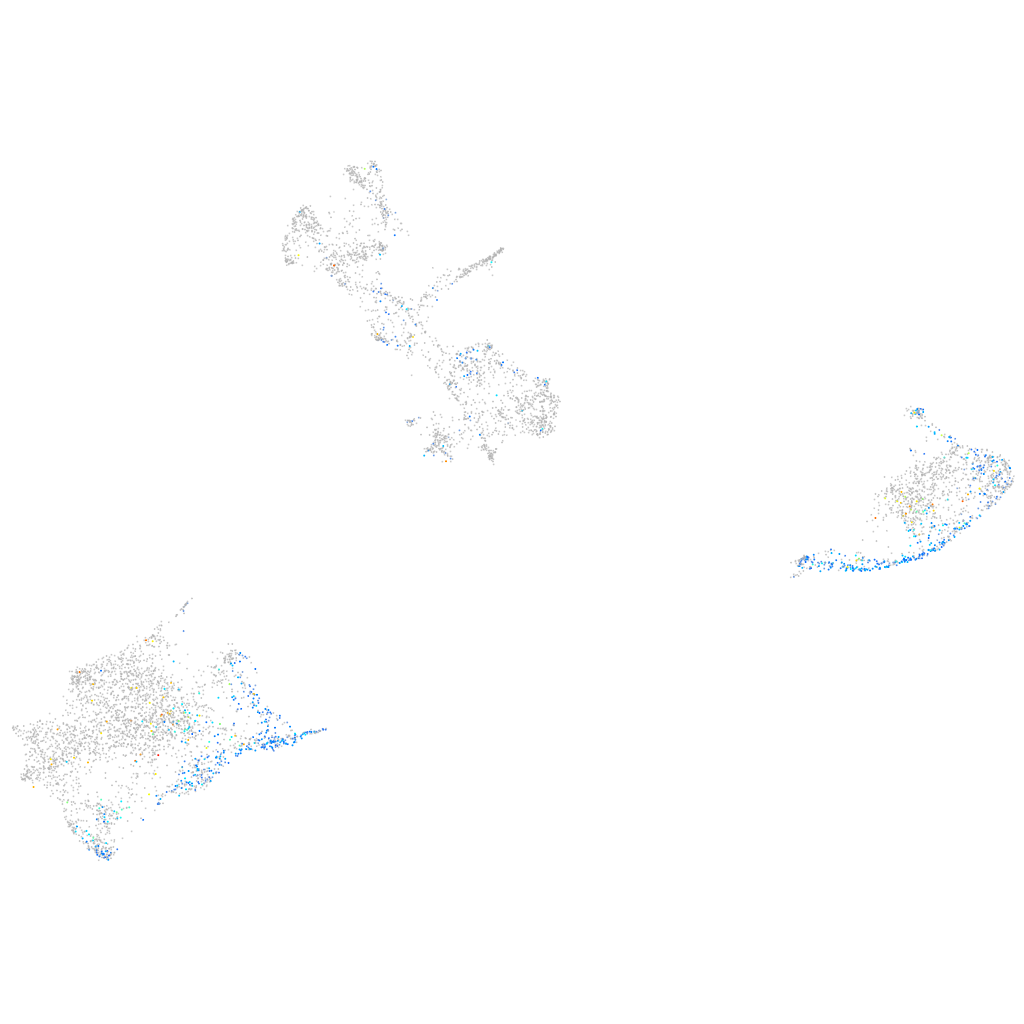
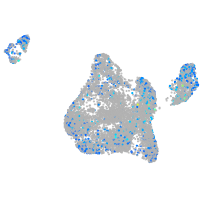
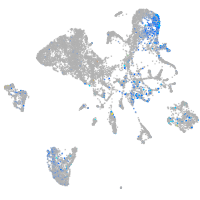
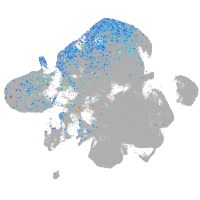
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| mitfa | 0.244 | pvalb1 | -0.128 |
| msx1b | 0.204 | pvalb2 | -0.127 |
| tfap2e | 0.199 | defbl1 | -0.114 |
| tmem243b | 0.195 | apoda.1 | -0.112 |
| rabl6b | 0.194 | mdkb | -0.111 |
| qdpra | 0.194 | ptmaa | -0.109 |
| xbp1 | 0.189 | lypc | -0.105 |
| tspan36 | 0.186 | gpnmb | -0.103 |
| slc37a2 | 0.184 | si:ch211-256m1.8 | -0.103 |
| ctsc | 0.184 | actc1b | -0.100 |
| XLOC-001964 | 0.181 | hbbe1.3 | -0.099 |
| phlda1 | 0.180 | si:dkey-197i20.6 | -0.096 |
| crestin | 0.179 | cdh11 | -0.092 |
| ctsba | 0.178 | unm-sa821 | -0.091 |
| pepd | 0.177 | hbae3 | -0.088 |
| atp6ap1b | 0.175 | s100v2 | -0.087 |
| syngr1a | 0.174 | gpm6aa | -0.085 |
| tmem243a | 0.174 | akap12a | -0.085 |
| C12orf75 | 0.173 | ponzr1 | -0.084 |
| slc3a2a | 0.173 | tuba1c | -0.082 |
| lman2 | 0.172 | cx43 | -0.081 |
| rgs2 | 0.171 | prx | -0.081 |
| cdh1 | 0.170 | si:ch73-89b15.3 | -0.081 |
| pah | 0.170 | tmsb | -0.080 |
| canx | 0.170 | plecb | -0.080 |
| sypl2b | 0.170 | slc25a36a | -0.079 |
| slc30a7 | 0.169 | hbae1.1 | -0.079 |
| tyr | 0.169 | alx4a | -0.079 |
| glb1l | 0.167 | tnni2a.4 | -0.078 |
| lamp1a | 0.166 | alx4b | -0.077 |
| SPAG9 | 0.164 | nova2 | -0.075 |
| naga | 0.164 | tmem179b | -0.075 |
| pcbd1 | 0.163 | sulf2b | -0.075 |
| pcdh10a | 0.161 | alx1 | -0.075 |
| aamp | 0.158 | guk1a | -0.074 |