acyl-CoA thioesterase 14
ZFIN 
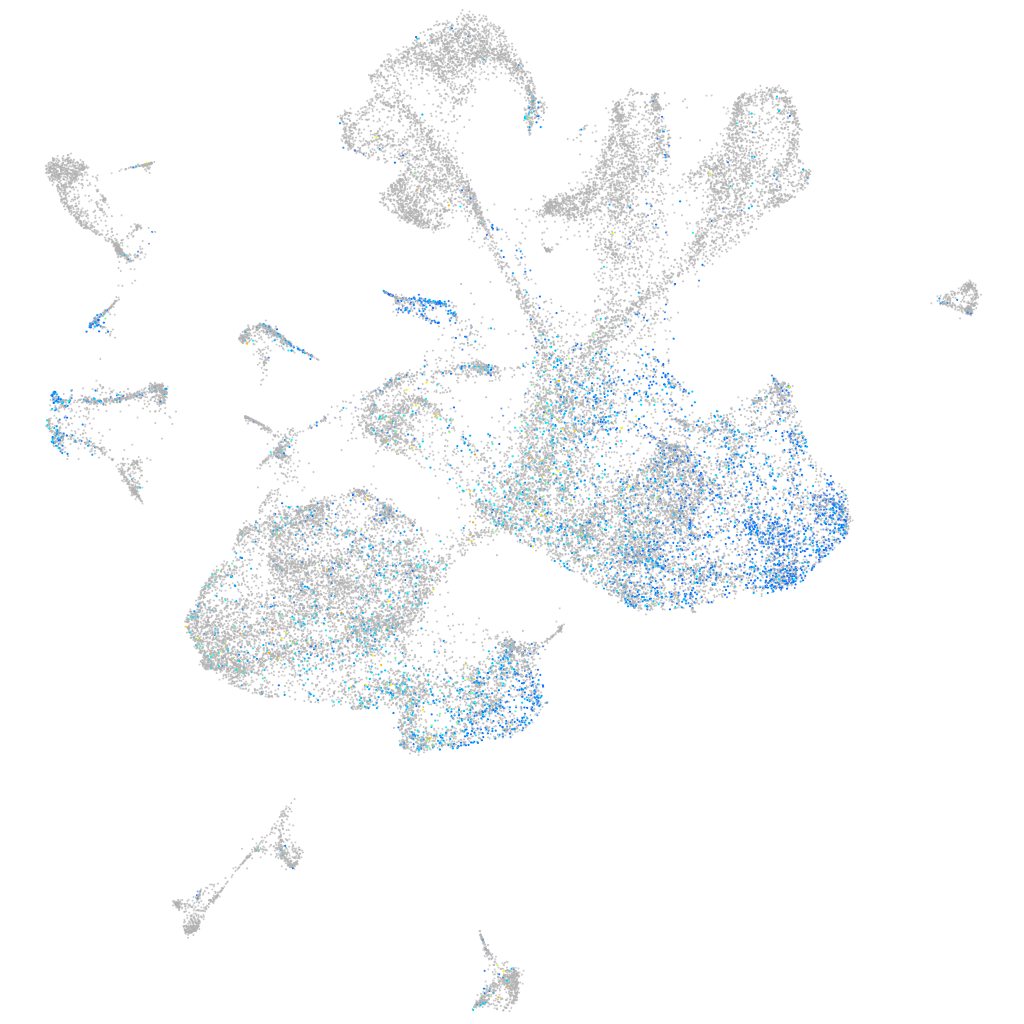
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| XLOC-003690 | 0.179 | gpm6ab | -0.150 |
| XLOC-003689 | 0.176 | stmn1b | -0.148 |
| cldn5a | 0.169 | elavl4 | -0.136 |
| hspb1 | 0.169 | ckbb | -0.135 |
| hmgb2a | 0.168 | gng3 | -0.135 |
| BX927258.1 | 0.160 | rtn1a | -0.127 |
| apela | 0.160 | vamp2 | -0.125 |
| id1 | 0.158 | sncb | -0.125 |
| cdx4 | 0.157 | stxbp1a | -0.123 |
| eef1a1l1 | 0.157 | stx1b | -0.123 |
| zgc:56493 | 0.156 | atp6v0cb | -0.123 |
| ranbp1 | 0.154 | myt1b | -0.120 |
| tspan7 | 0.153 | snap25a | -0.119 |
| her15.1 | 0.152 | rtn1b | -0.117 |
| her12 | 0.152 | rnasekb | -0.113 |
| sox3 | 0.152 | pvalb2 | -0.113 |
| cx43.4 | 0.151 | nsg2 | -0.112 |
| ran | 0.151 | cplx2 | -0.112 |
| CABZ01075068.1 | 0.150 | stmn2a | -0.112 |
| selenoh | 0.149 | pvalb1 | -0.111 |
| npm1a | 0.146 | ywhag2 | -0.111 |
| btg2 | 0.146 | si:dkeyp-75h12.5 | -0.110 |
| mcm7 | 0.145 | csdc2a | -0.109 |
| pcna | 0.144 | gap43 | -0.108 |
| dut | 0.141 | tmem59l | -0.107 |
| snrpb | 0.140 | ywhah | -0.107 |
| XLOC-032526 | 0.140 | syt2a | -0.106 |
| tpm4a | 0.139 | gapdhs | -0.105 |
| snrpf | 0.139 | calm1a | -0.105 |
| rps7 | 0.138 | rbfox1 | -0.104 |
| sox19a | 0.138 | elavl3 | -0.104 |
| sb:cb81 | 0.137 | aplp1 | -0.104 |
| nhp2 | 0.137 | dpysl3 | -0.103 |
| h2afvb | 0.137 | zgc:65894 | -0.102 |
| rpa2 | 0.137 | id4 | -0.101 |