acyl-CoA binding domain containing 5b
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature 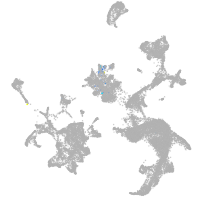 |
pronephros  |
neural |
spinal cord / glia |
eye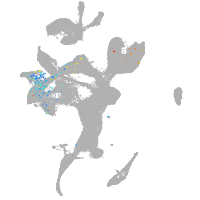 |
taste / olfactory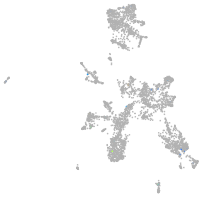 |
otic / lateral line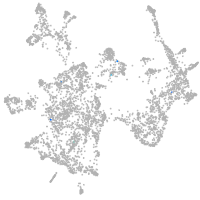 |
epidermis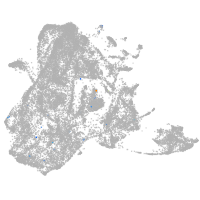 |
periderm |
fin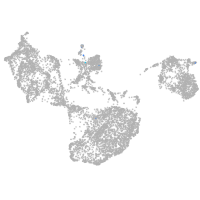 |
ionocytes / mucous |
pigment cells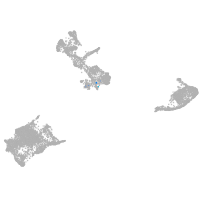 |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| CU469539.1 | 0.632 | rplp2l | -0.047 |
| scn12aa | 0.497 | rpl9 | -0.040 |
| si:ch211-266k22.6 | 0.444 | rps13 | -0.039 |
| CR848741.1 | 0.433 | rpl35 | -0.038 |
| CABZ01072406.1 | 0.429 | rps4x | -0.037 |
| calhm3 | 0.429 | tspan36 | -0.033 |
| FO834800.2 | 0.429 | rpl34 | -0.032 |
| crhr2.2 | 0.429 | rplp0 | -0.030 |
| endou | 0.372 | si:dkey-151g10.6 | -0.030 |
| RNF157 | 0.370 | tpt1 | -0.030 |
| wasf3a | 0.326 | rpl36 | -0.028 |
| npffr1 | 0.324 | rpl38 | -0.028 |
| XLOC-032843 | 0.311 | rplp1 | -0.027 |
| ripply3 | 0.304 | zgc:56493 | -0.027 |
| cdkn2c | 0.303 | rpl7 | -0.027 |
| tnnt1 | 0.303 | eef1g | -0.026 |
| zp2l2 | 0.302 | smim29 | -0.025 |
| itpr1b | 0.297 | rpl5b | -0.024 |
| nr1d4a | 0.290 | rps10 | -0.024 |
| tmprss15 | 0.290 | rpl18a | -0.024 |
| trim46b | 0.288 | eef1b2 | -0.024 |
| sim1b | 0.284 | rps19 | -0.024 |
| CABZ01060030.1 | 0.278 | rps7 | -0.024 |
| arhgap20 | 0.278 | gpr143 | -0.024 |
| CU695222.2 | 0.270 | atox1 | -0.024 |
| si:dkey-26h11.2 | 0.270 | gmps | -0.023 |
| atrnl1a | 0.269 | rps25 | -0.023 |
| myo18aa | 0.261 | rpl7a | -0.023 |
| snx8b | 0.236 | rpl18 | -0.022 |
| ppp1r1b | 0.230 | slc2a15a | -0.022 |
| si:ch73-305o9.3 | 0.229 | rpl4 | -0.021 |
| camkvl | 0.229 | rpl27a | -0.021 |
| mir132-2 | 0.229 | ctsd | -0.021 |
| qpctlb | 0.227 | rps20 | -0.021 |
| tacr2 | 0.225 | rpsa | -0.021 |