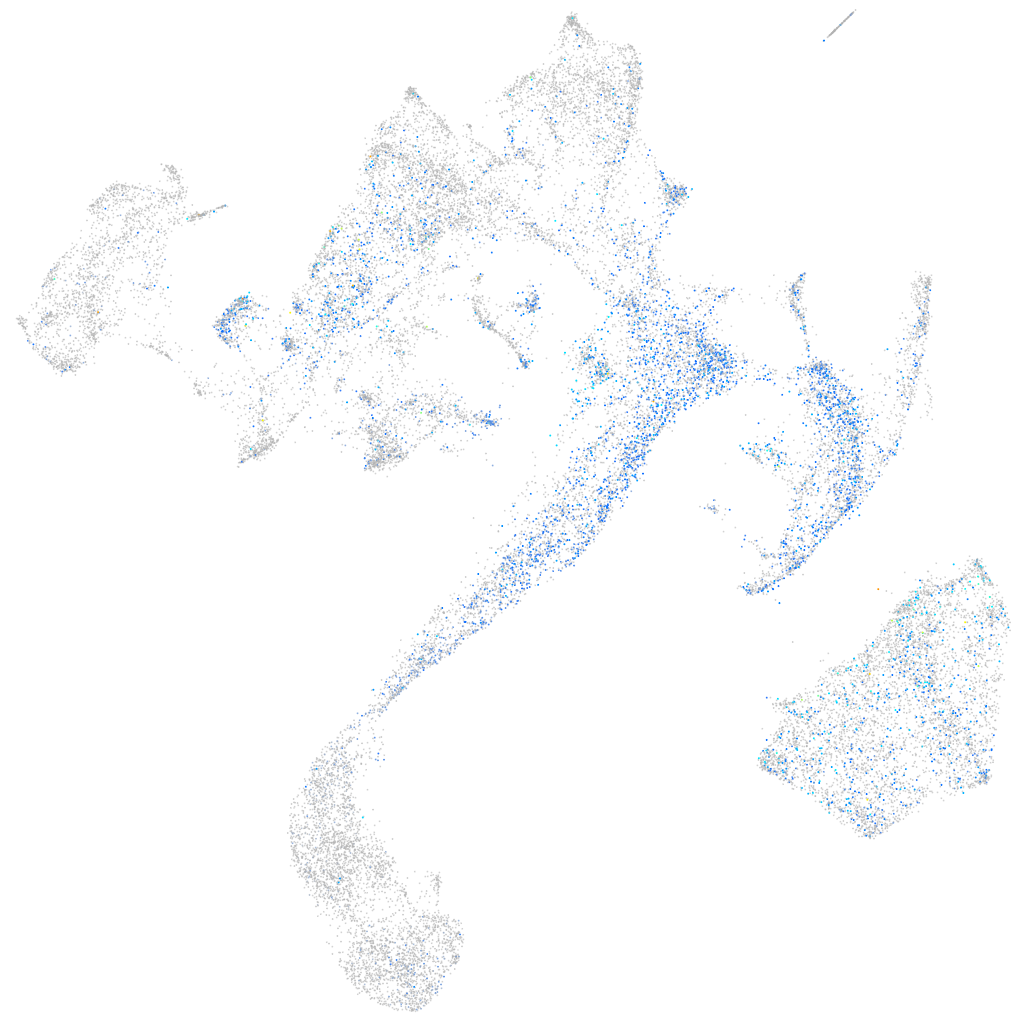
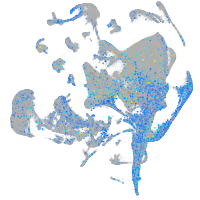
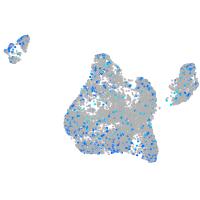
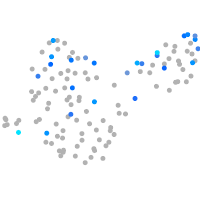
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| snu13b | 0.229 | si:dkey-16p21.8 | -0.151 |
| npm1a | 0.226 | ckma | -0.145 |
| nhp2 | 0.218 | eno3 | -0.145 |
| nop10 | 0.209 | actn3a | -0.143 |
| rpl7l1 | 0.209 | neb | -0.142 |
| cnbpa | 0.207 | pvalb1 | -0.140 |
| setb | 0.205 | smyd1a | -0.139 |
| nop56 | 0.200 | gapdh | -0.138 |
| nop58 | 0.200 | si:ch73-367p23.2 | -0.138 |
| tsr2 | 0.199 | bhmt | -0.138 |
| ddx39ab | 0.198 | ckmb | -0.137 |
| cdkn2aipnl | 0.197 | tnnc2 | -0.136 |
| snrpf | 0.196 | si:ch211-266g18.10 | -0.135 |
| bxdc2 | 0.195 | myom1a | -0.135 |
| tomm20a | 0.194 | ak1 | -0.135 |
| ssb | 0.194 | actn3b | -0.134 |
| snrpd2 | 0.192 | actc1b | -0.134 |
| bysl | 0.191 | XLOC-025819 | -0.134 |
| emg1 | 0.190 | pgam2 | -0.134 |
| nip7 | 0.189 | pvalb2 | -0.133 |
| ran | 0.189 | pmp22b | -0.133 |
| mrto4 | 0.189 | prx | -0.133 |
| nifk | 0.189 | tmod4 | -0.132 |
| snrpe | 0.189 | ndrg2 | -0.132 |
| rcl1 | 0.188 | casq2 | -0.132 |
| ptges3b | 0.188 | ldb3b | -0.131 |
| prmt1 | 0.186 | XLOC-001975 | -0.129 |
| snrpd1 | 0.185 | nme2b.2 | -0.129 |
| npm3 | 0.185 | XLOC-005350 | -0.128 |
| dkc1 | 0.185 | atp2a1 | -0.128 |
| ncl | 0.184 | myom1b | -0.128 |
| hnrnpabb | 0.181 | sgcd | -0.128 |
| snrpb | 0.181 | XLOC-006515 | -0.127 |
| ebna1bp2 | 0.178 | tnnt3b | -0.127 |
| tma16 | 0.178 | pkmb | -0.126 |