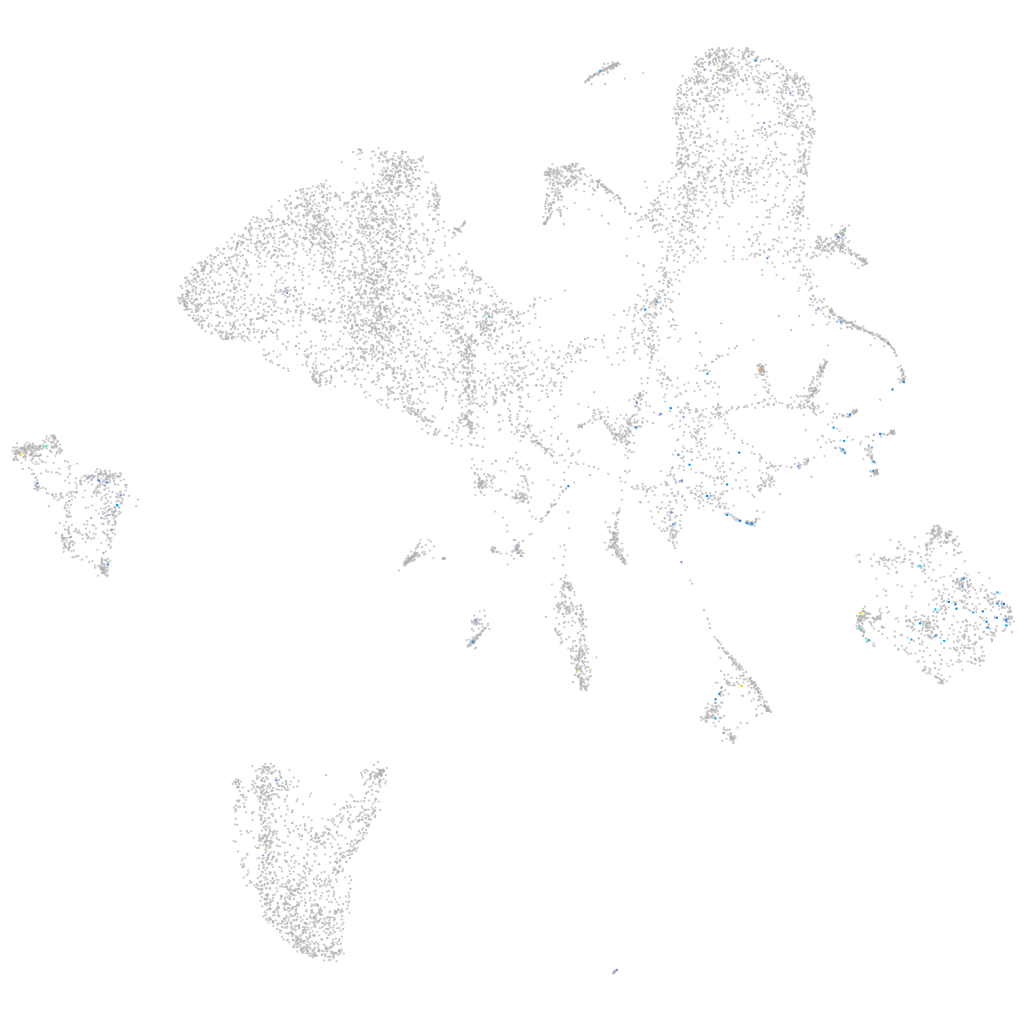
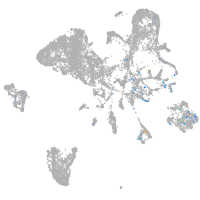
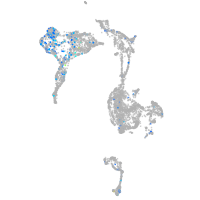
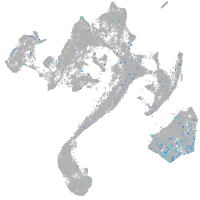
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| znf1085 | 0.224 | ahcy | -0.085 |
| CU914622.1 | 0.224 | gamt | -0.082 |
| AL953886.2 | 0.181 | gapdh | -0.081 |
| prlh2 | 0.170 | gatm | -0.074 |
| tlr22 | 0.163 | bhmt | -0.074 |
| LOC101884135 | 0.145 | eno3 | -0.072 |
| FP102783.1 | 0.140 | mat1a | -0.071 |
| CR450848.1 | 0.139 | sod1 | -0.068 |
| LOC110440222 | 0.133 | apoa1b | -0.066 |
| XLOC-011190 | 0.132 | gpx4a | -0.066 |
| mitfb | 0.130 | agxtb | -0.064 |
| LOC103910403 | 0.129 | pklr | -0.064 |
| LOC110438235 | 0.129 | suclg1 | -0.063 |
| LOC103909615 | 0.128 | fbp1b | -0.063 |
| LOC108180138 | 0.125 | scp2a | -0.063 |
| filip1a | 0.124 | glud1b | -0.062 |
| lgals17 | 0.124 | apoa4b.1 | -0.061 |
| pik3ip1 | 0.118 | apoc2 | -0.061 |
| si:dkey-100n19.2 | 0.117 | atp5f1b | -0.061 |
| elob | 0.117 | agxta | -0.061 |
| shroom3 | 0.116 | apoa2 | -0.060 |
| ctbs | 0.113 | prdx2 | -0.059 |
| LOC101885980 | 0.112 | aldh6a1 | -0.059 |
| st8sia4 | 0.111 | mt-atp6 | -0.058 |
| chgb | 0.109 | ckba | -0.058 |
| CABZ01046427.1 | 0.109 | grhprb | -0.058 |
| mon1ba | 0.106 | cx32.3 | -0.058 |
| zgc:158270 | 0.105 | afp4 | -0.058 |
| gpr84 | 0.104 | nipsnap3a | -0.058 |
| XLOC-043113 | 0.103 | abat | -0.057 |
| pkd1l1 | 0.102 | ces2 | -0.057 |
| apba2a | 0.101 | gstt1a | -0.057 |
| ftr76 | 0.100 | fabp10a | -0.057 |
| AL645792.1 | 0.099 | mdh1aa | -0.057 |
| LO018363.1 | 0.099 | ttc36 | -0.056 |