"zinc finger (CCCH type), RNA-binding motif and serine/arginine rich 2"
ZFIN 
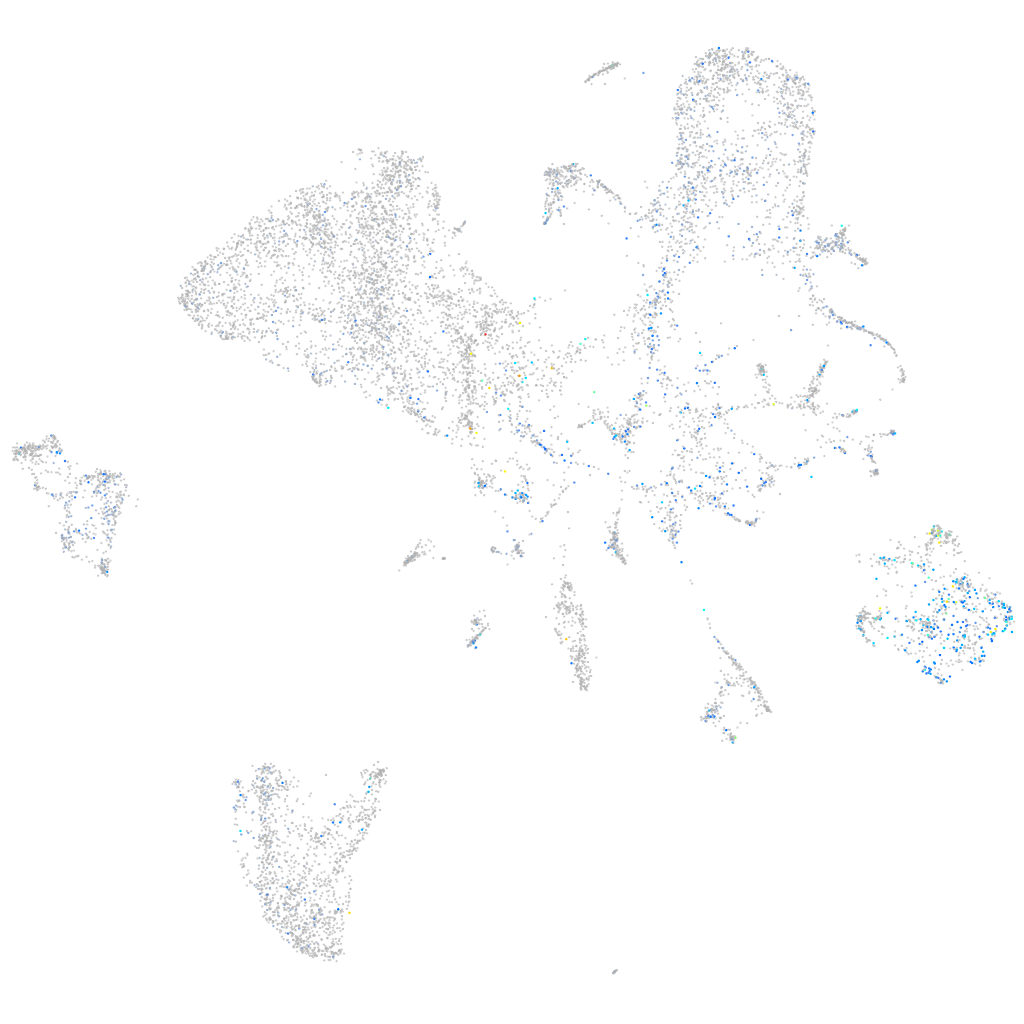
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| sox32 | 0.251 | rps10 | -0.180 |
| marcksb | 0.249 | rpl37 | -0.175 |
| nucks1a | 0.249 | gamt | -0.175 |
| hnrnpa1b | 0.248 | zgc:92744 | -0.173 |
| top1l | 0.247 | zgc:114188 | -0.164 |
| brd3a | 0.246 | ahcy | -0.163 |
| cxcr4a | 0.244 | gapdh | -0.158 |
| acin1a | 0.244 | bhmt | -0.154 |
| marcksl1b | 0.243 | gatm | -0.153 |
| ppig | 0.242 | rps17 | -0.151 |
| ncl | 0.242 | apoa1b | -0.147 |
| hnrnpa1a | 0.239 | gpx4a | -0.144 |
| tbx16 | 0.238 | mat1a | -0.143 |
| ilf3b | 0.238 | pnp4b | -0.142 |
| si:ch211-152c2.3 | 0.238 | agxtb | -0.142 |
| hnrnpub | 0.237 | apoa2 | -0.142 |
| akap12b | 0.237 | apoa4b.1 | -0.139 |
| smc1al | 0.236 | pklr | -0.138 |
| hmga1a | 0.236 | fbp1b | -0.138 |
| snrnp70 | 0.235 | suclg1 | -0.137 |
| snrpa | 0.235 | sod1 | -0.137 |
| seta | 0.233 | eno3 | -0.137 |
| srrm2 | 0.233 | tpi1b | -0.135 |
| lig1 | 0.232 | grhprb | -0.134 |
| hspb1 | 0.232 | nupr1b | -0.133 |
| prpf40a | 0.232 | atp5if1b | -0.133 |
| si:dkey-66i24.9 | 0.231 | afp4 | -0.132 |
| hnrnpm | 0.231 | ces2 | -0.132 |
| sp5l | 0.231 | nme2b.1 | -0.132 |
| syncrip | 0.230 | pgm1 | -0.131 |
| LOC100008030 | 0.230 | ckba | -0.131 |
| cx43.4 | 0.229 | serpina1 | -0.131 |
| zgc:110425 | 0.229 | rbp4 | -0.130 |
| chaf1a | 0.229 | fabp3 | -0.130 |
| snrpd3l | 0.228 | fetub | -0.129 |