zinc finger protein 407
ZFIN 
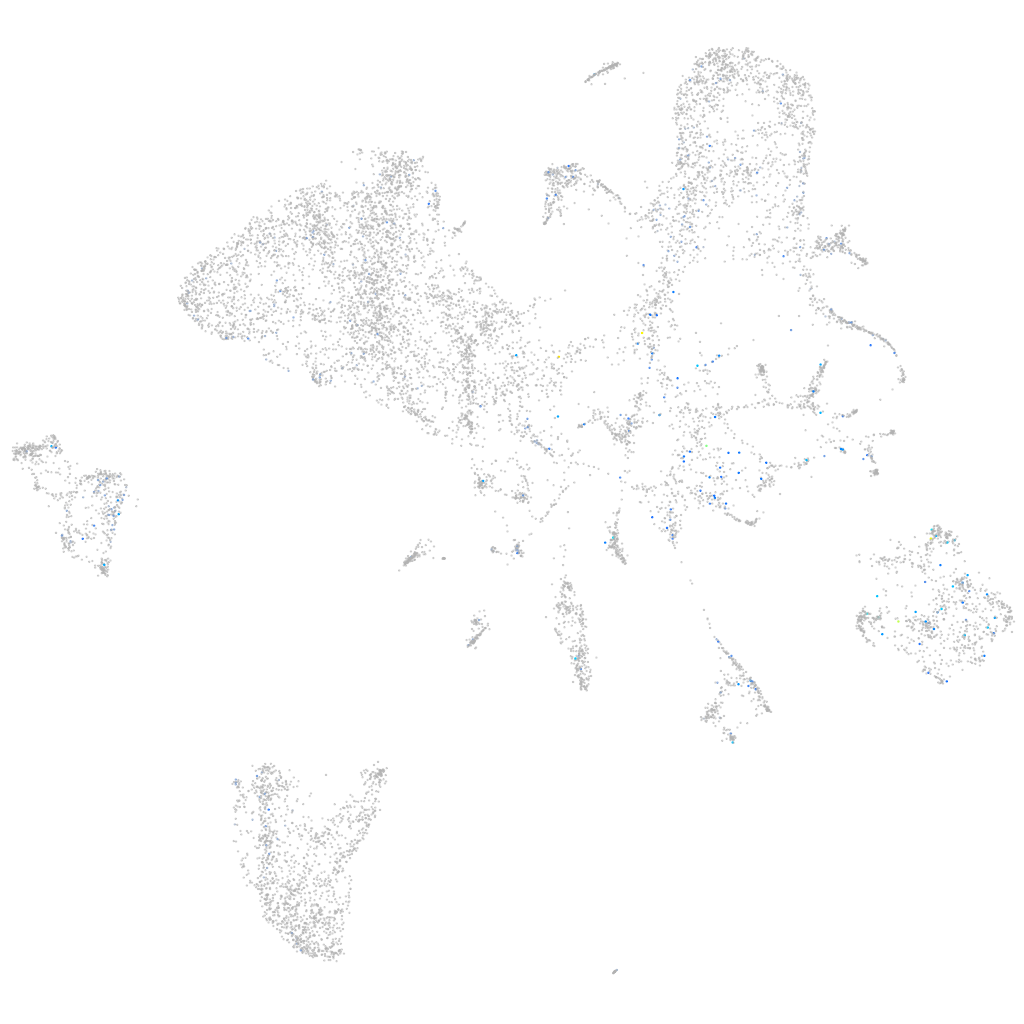
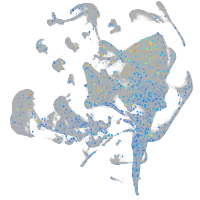
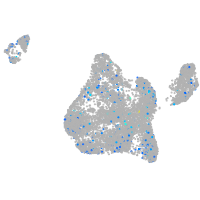
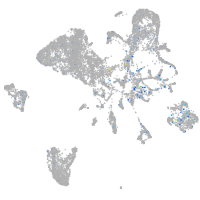
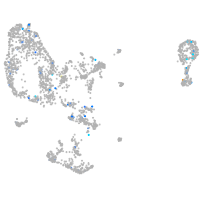
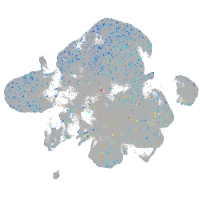
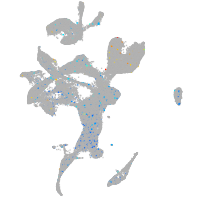
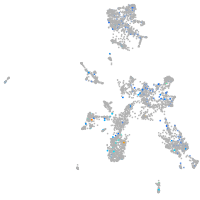
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| BX957276.1 | 0.167 | gamt | -0.088 |
| BX322665.1 | 0.160 | gatm | -0.086 |
| LOC101886187 | 0.157 | gapdh | -0.084 |
| LOC103909803 | 0.154 | bhmt | -0.081 |
| BX465198.1 | 0.147 | apoa1b | -0.074 |
| zgc:165453 | 0.144 | mat1a | -0.073 |
| CT027759.1 | 0.144 | gpx4a | -0.072 |
| LOC101885165 | 0.142 | pnp4b | -0.070 |
| BX005442.1 | 0.132 | apoa2 | -0.069 |
| or112-1 | 0.131 | agxtb | -0.069 |
| si:dkeyp-82b4.3 | 0.127 | ahcy | -0.068 |
| CR855318.1 | 0.118 | apoa4b.1 | -0.066 |
| LOC108180723 | 0.118 | dap | -0.066 |
| LOC110437790 | 0.117 | fbp1b | -0.065 |
| nrp1b | 0.115 | nupr1b | -0.065 |
| CR407590.2 | 0.113 | rbp4 | -0.064 |
| si:dkey-23f9.8 | 0.112 | scp2a | -0.064 |
| FP102888.2 | 0.111 | afp4 | -0.064 |
| CT025776.1 | 0.110 | grhprb | -0.064 |
| cirbpa | 0.108 | abat | -0.063 |
| cbx3a | 0.106 | fabp10a | -0.062 |
| smc1al | 0.106 | apom | -0.062 |
| ncam3 | 0.106 | apoc2 | -0.062 |
| nucks1a | 0.106 | rbp2b | -0.061 |
| si:ch211-286b4.4 | 0.106 | zgc:123103 | -0.060 |
| khdrbs1a | 0.105 | zgc:92744 | -0.060 |
| si:ch211-222l21.1 | 0.105 | tfa | -0.060 |
| LOC101887111 | 0.104 | serpina1 | -0.059 |
| anp32b | 0.104 | eno3 | -0.059 |
| hmga1a | 0.104 | pklr | -0.059 |
| trim35-7 | 0.103 | fetub | -0.059 |
| nono | 0.102 | ttr | -0.059 |
| anp32a | 0.102 | pgm1 | -0.058 |
| uckl1a | 0.101 | ces2 | -0.058 |
| hmgn6 | 0.100 | apobb.1 | -0.058 |