zinc finger protein 1146
ZFIN 
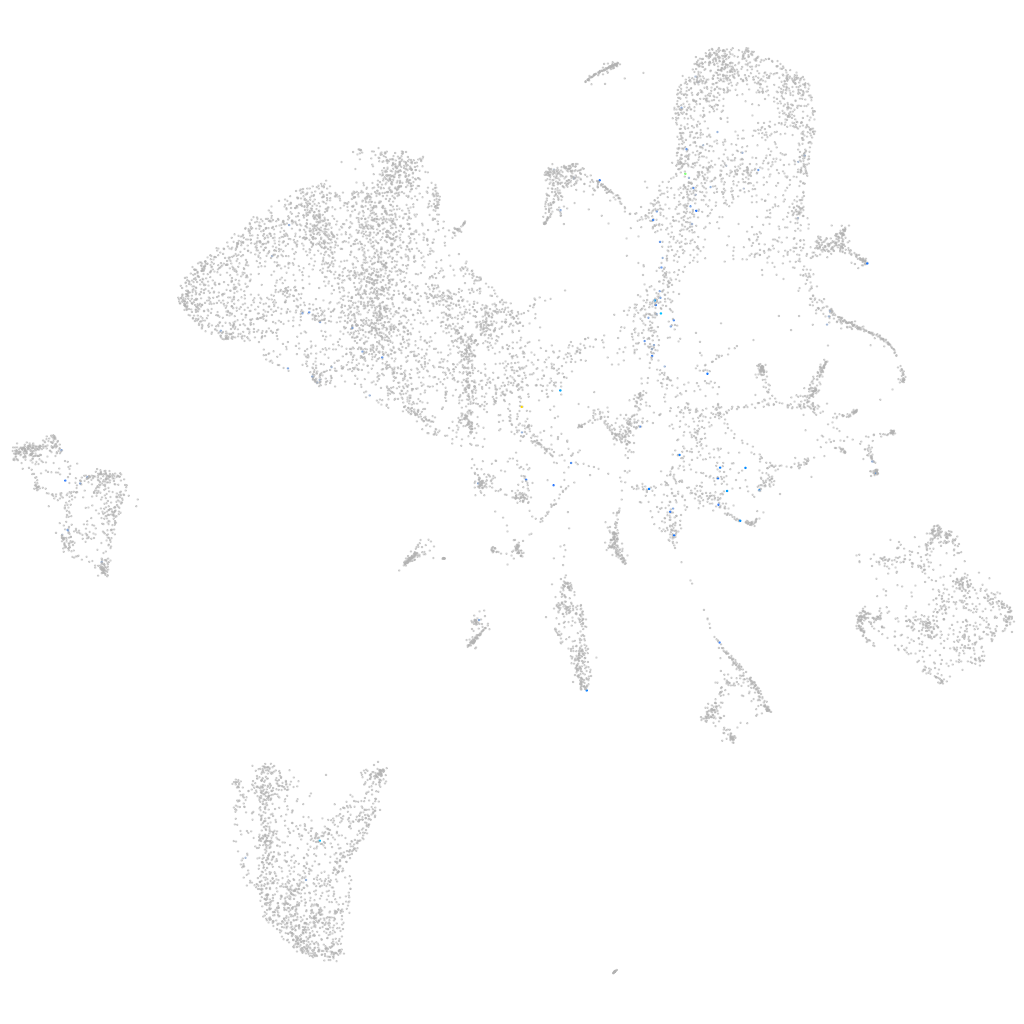
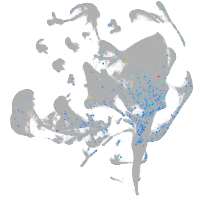
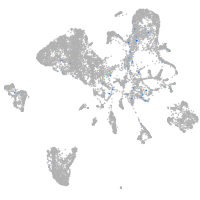
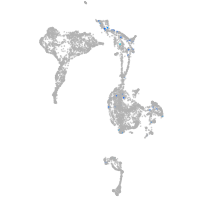
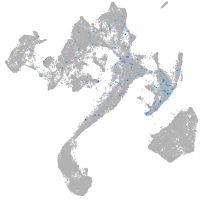
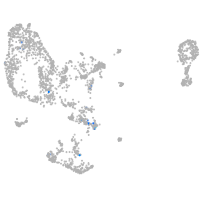
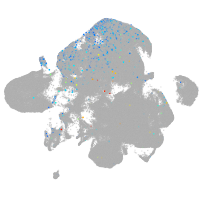
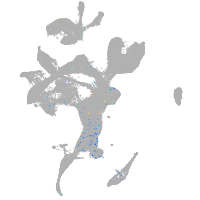
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| pimr197 | 0.388 | pnp4b | -0.043 |
| LOC101886261 | 0.295 | apoc1 | -0.042 |
| LOC101884462 | 0.279 | agxtb | -0.040 |
| si:dkey-7j22.4 | 0.270 | gamt | -0.039 |
| cart2 | 0.243 | ckba | -0.039 |
| gria4b | 0.236 | bhmt | -0.039 |
| mrc1a | 0.201 | gatm | -0.038 |
| si:dkeyp-44a8.4 | 0.192 | tfa | -0.036 |
| CABZ01084942.1 | 0.188 | rbp4 | -0.036 |
| psmb9a | 0.163 | zgc:123103 | -0.036 |
| LOC101883330 | 0.159 | fabp10a | -0.036 |
| CT737131.1 | 0.158 | grhprb | -0.036 |
| LOC100000752 | 0.158 | gapdh | -0.036 |
| XLOC-043650 | 0.143 | serpina1 | -0.036 |
| LOC103910169 | 0.139 | ces2 | -0.035 |
| CU329666.1 | 0.138 | fetub | -0.035 |
| XLOC-034224 | 0.134 | serpina1l | -0.035 |
| CR626887.1 | 0.126 | cyp2n13 | -0.035 |
| si:ch211-76l23.7 | 0.126 | rbp2b | -0.035 |
| CABZ01046427.1 | 0.125 | hpda | -0.034 |
| si:dkey-27n6.1 | 0.120 | fgg | -0.034 |
| BX546459.1 | 0.118 | pck1 | -0.034 |
| chst12b | 0.117 | gda | -0.034 |
| lzts2b | 0.117 | apom | -0.034 |
| CU013548.1 | 0.114 | fgb | -0.034 |
| zgc:152857 | 0.113 | gc | -0.033 |
| BX511224.1 | 0.111 | mat1a | -0.033 |
| rims3 | 0.108 | comtd1 | -0.033 |
| kcna1a | 0.106 | ucp1 | -0.033 |
| si:ch211-214b16.3 | 0.104 | cyp2ad2 | -0.033 |
| brat1 | 0.102 | cp | -0.033 |
| TIMM22 | 0.101 | tdo2a | -0.033 |
| loxl5b | 0.100 | abat | -0.033 |
| CABZ01059120.1 | 0.100 | uox | -0.033 |
| LOC108180355 | 0.097 | c9 | -0.032 |