zinc finger protein 1134
ZFIN 
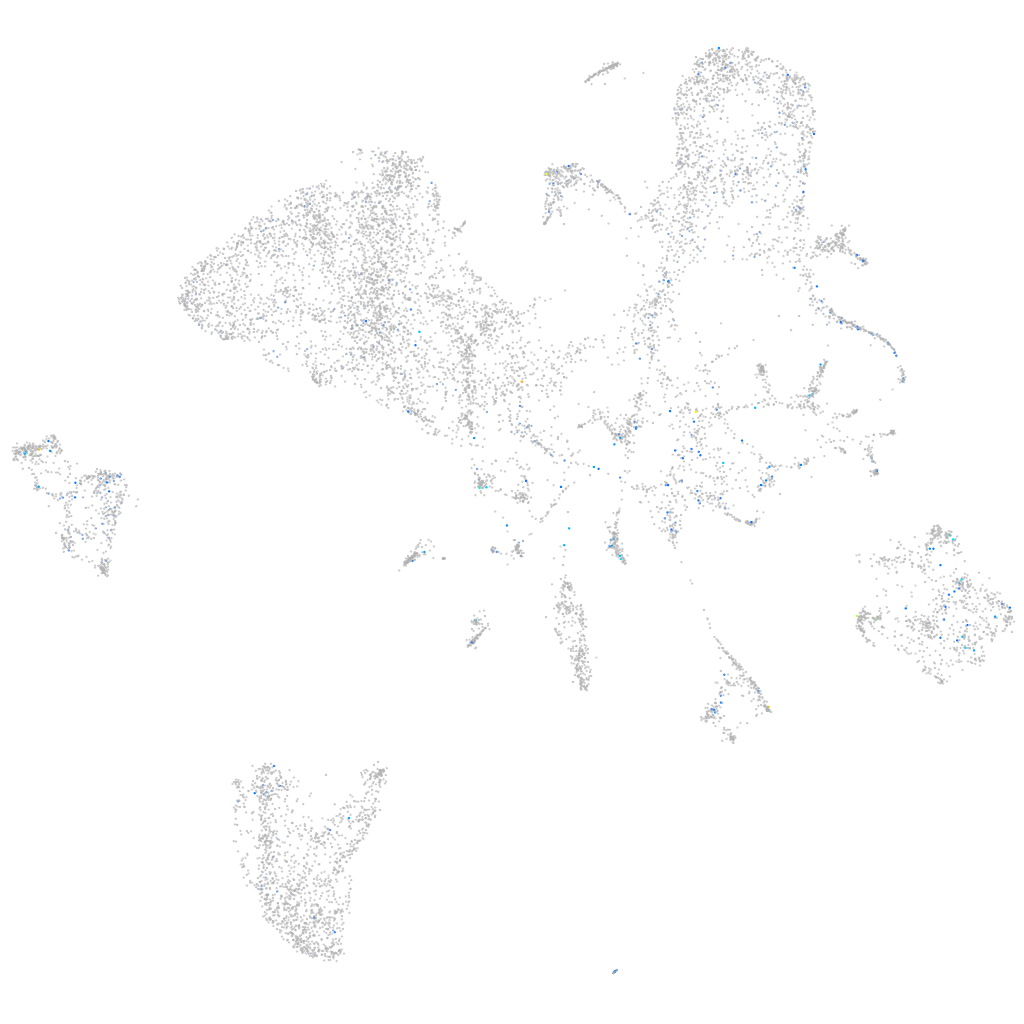
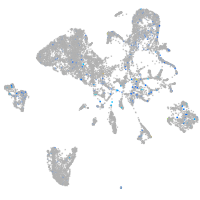
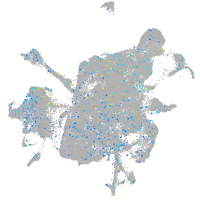
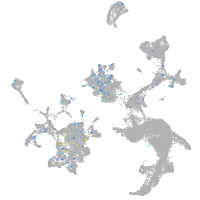
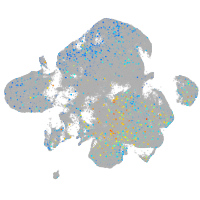
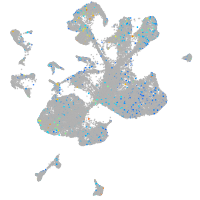
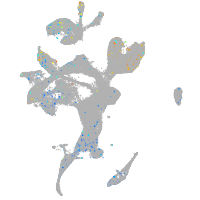
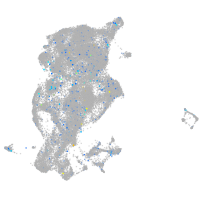
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| FQ790367.1 | 0.169 | bhmt | -0.087 |
| adra1ba | 0.166 | gamt | -0.086 |
| XLOC-014193 | 0.148 | gatm | -0.084 |
| si:ch1073-268j14.1 | 0.133 | ahcy | -0.080 |
| fam113 | 0.123 | gapdh | -0.079 |
| LOC110440119 | 0.122 | agxtb | -0.076 |
| XLOC-007246 | 0.117 | mat1a | -0.074 |
| LOC110438082 | 0.115 | apoa1b | -0.074 |
| XLOC-026848 | 0.109 | grhprb | -0.073 |
| themis | 0.108 | apoa2 | -0.070 |
| CR381540.3 | 0.108 | aqp12 | -0.070 |
| LOC100537734 | 0.108 | afp4 | -0.070 |
| CR626886.2 | 0.106 | apom | -0.069 |
| TMEM196 | 0.103 | zgc:123103 | -0.067 |
| LOC562770 | 0.101 | serpina1 | -0.067 |
| CT990625.1 | 0.100 | gpx4a | -0.067 |
| prok1 | 0.098 | serpina1l | -0.067 |
| LOC100535286 | 0.098 | tfa | -0.067 |
| CABZ01088229.1 | 0.098 | ces2 | -0.067 |
| pld3 | 0.097 | fetub | -0.066 |
| ccdc80 | 0.097 | rbp2b | -0.066 |
| CU019562.3 | 0.097 | kng1 | -0.066 |
| zgc:123299 | 0.097 | hao1 | -0.065 |
| si:ch211-203k16.3 | 0.096 | fabp10a | -0.065 |
| ubc | 0.095 | fgg | -0.065 |
| zgc:92313 | 0.095 | LOC110437731 | -0.064 |
| ggact.2 | 0.094 | aldh7a1 | -0.064 |
| wu:fb55g09 | 0.094 | abat | -0.064 |
| CABZ01077220.1 | 0.094 | suclg1 | -0.064 |
| GPR45 | 0.093 | fgb | -0.064 |
| cavin2b | 0.093 | agxta | -0.064 |
| ftr87 | 0.091 | c9 | -0.063 |
| XLOC-033528 | 0.091 | ambp | -0.063 |
| ier5 | 0.091 | ttc36 | -0.063 |
| loxl3a | 0.090 | apoa4b.1 | -0.063 |