zinc finger protein 1010
ZFIN 
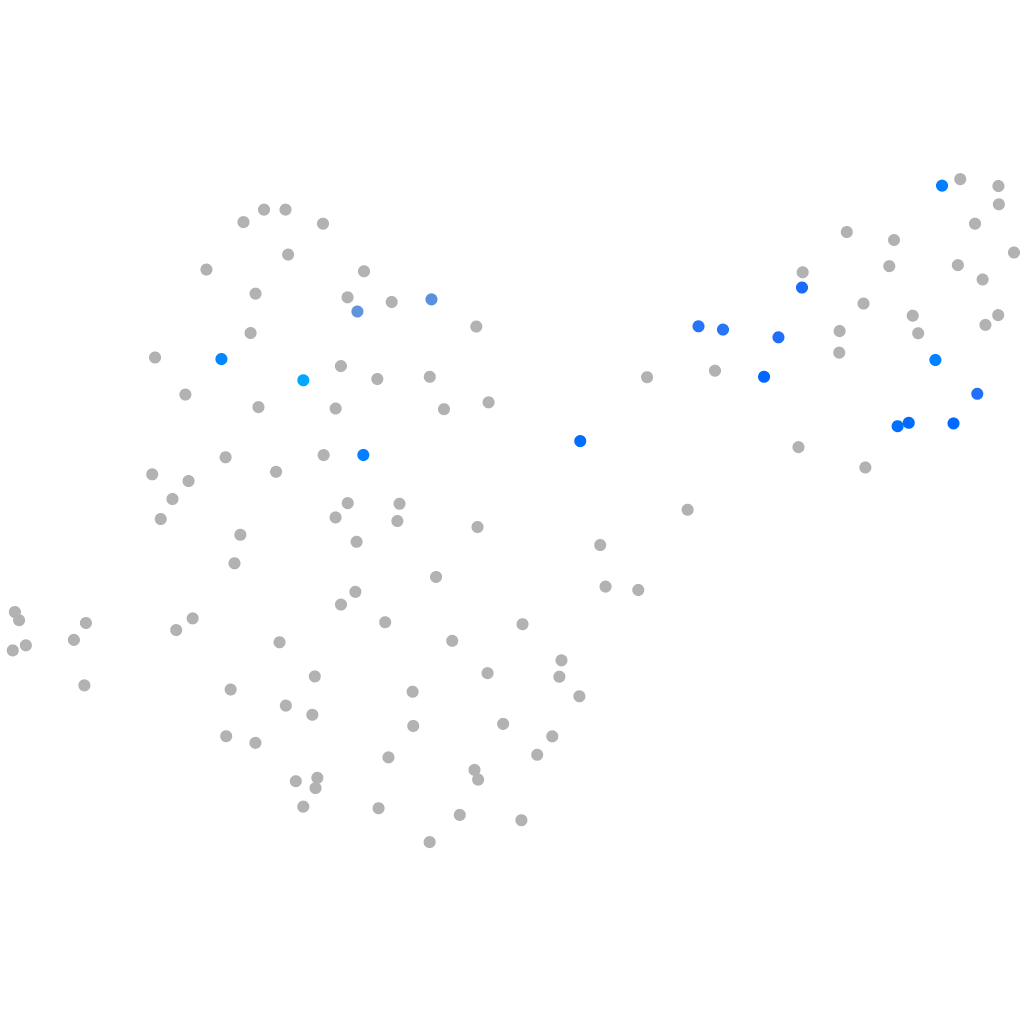
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| bcor | 0.498 | sde2 | -0.225 |
| adra1ba | 0.493 | ISCU | -0.215 |
| fhl3a | 0.485 | snrpb2 | -0.206 |
| pot1 | 0.484 | h1m | -0.200 |
| celf2 | 0.468 | nop58 | -0.199 |
| sdr16c5a | 0.467 | phpt1 | -0.195 |
| fgfbp1b | 0.449 | si:ch211-51e12.7 | -0.193 |
| hps1 | 0.449 | ppig | -0.192 |
| shc1 | 0.448 | elavl1a | -0.191 |
| mir135b | 0.441 | xpo6 | -0.188 |
| BX571811.2 | 0.440 | dnaja2a | -0.186 |
| fdx1b | 0.439 | vox | -0.183 |
| stim1a | 0.436 | arih2 | -0.180 |
| b3gnt2b | 0.429 | srrt | -0.179 |
| rnf26 | 0.426 | thap7 | -0.178 |
| dab2ipa | 0.426 | mis12 | -0.172 |
| slc7a1 | 0.423 | NC-002333.4 | -0.171 |
| si:dkeyp-26a9.2 | 0.422 | emc9 | -0.170 |
| BX511136.1 | 0.420 | ss18 | -0.170 |
| LRRTM4 | 0.420 | rrp1 | -0.167 |
| efnb3b | 0.419 | COX7A2 | -0.167 |
| her15.1 | 0.418 | nasp | -0.166 |
| smarcd2 | 0.418 | rhoca | -0.164 |
| ap1ar | 0.416 | msl1a | -0.163 |
| XLOC-042971 | 0.416 | si:ch211-152c2.3 | -0.163 |
| si:ch211-136m16.8 | 0.416 | nnr | -0.162 |
| si:ch211-262e15.1 | 0.415 | fscn1a | -0.161 |
| kcnip3b | 0.415 | knl1 | -0.161 |
| dacha | 0.414 | ccnb1 | -0.160 |
| pigs | 0.413 | zgc:56676 | -0.159 |
| XLOC-044244 | 0.412 | ccna2 | -0.158 |
| si:ch73-194h10.2 | 0.410 | si:dkeyp-20e4.8 | -0.158 |
| cacna1aa | 0.407 | bokb | -0.157 |
| pld1a | 0.407 | exosc9 | -0.157 |
| sc5d | 0.404 | mis18bp1 | -0.155 |