zgc:63470
ZFIN 
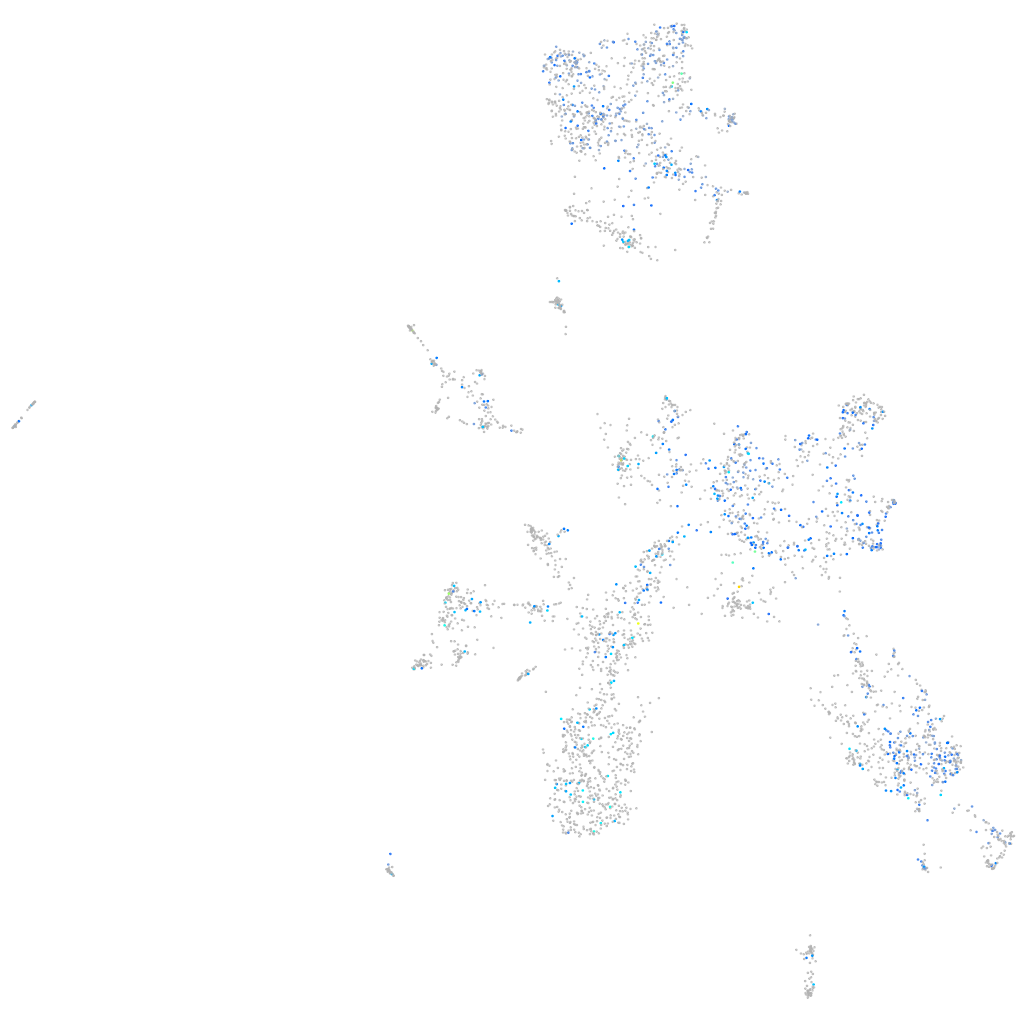
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| khdrbs1a | 0.153 | adgrl3.1 | -0.072 |
| hnrnpa0l | 0.148 | stx1b | -0.069 |
| snrpf | 0.140 | mt-co1 | -0.069 |
| hmgb2b | 0.138 | txn | -0.068 |
| ranbp1 | 0.138 | aldocb | -0.068 |
| ybx1 | 0.138 | COX3 | -0.067 |
| snrpb | 0.136 | camk2n1a | -0.063 |
| ran | 0.133 | NC-002333.17 | -0.062 |
| si:ch211-288g17.3 | 0.132 | sprn2 | -0.059 |
| hnrnpa0b | 0.128 | atp6v0cb | -0.058 |
| eef1b2 | 0.127 | slc6a17 | -0.058 |
| hnrnpabb | 0.127 | rd3 | -0.058 |
| ddx39aa | 0.127 | NC-002333.4 | -0.057 |
| tomm5 | 0.127 | atp6v0a1b | -0.057 |
| tcp1 | 0.126 | tnks1bp1 | -0.057 |
| cnbpa | 0.126 | pcloa | -0.056 |
| cct3 | 0.125 | rapgef2 | -0.056 |
| dut | 0.125 | syt1a | -0.055 |
| eif4a1a | 0.125 | CR383676.1 | -0.055 |
| seta | 0.123 | hbbe1.1 | -0.052 |
| cirbpa | 0.123 | bin1a | -0.052 |
| syncrip | 0.122 | mt-co2 | -0.052 |
| h2afvb | 0.122 | nfe2l2a | -0.051 |
| srsf2a | 0.121 | tuba2 | -0.051 |
| sp8a | 0.121 | mtmr14 | -0.051 |
| rpl10a | 0.121 | pvalb2 | -0.051 |
| snrpd1 | 0.121 | eef1a1a | -0.051 |
| stmn1a | 0.120 | si:ch73-60h1.1 | -0.050 |
| rplp0 | 0.120 | fstl5 | -0.050 |
| rbmx | 0.120 | tbcelb | -0.050 |
| snrpd2 | 0.119 | rnf41l | -0.050 |
| nudt21 | 0.119 | camk1gb | -0.050 |
| her15.1 | 0.118 | bdnf | -0.050 |
| eif4ebp1 | 0.118 | jakmip1 | -0.049 |
| snrpg | 0.118 | wu:fb18f06 | -0.049 |