zgc:55262
ZFIN 
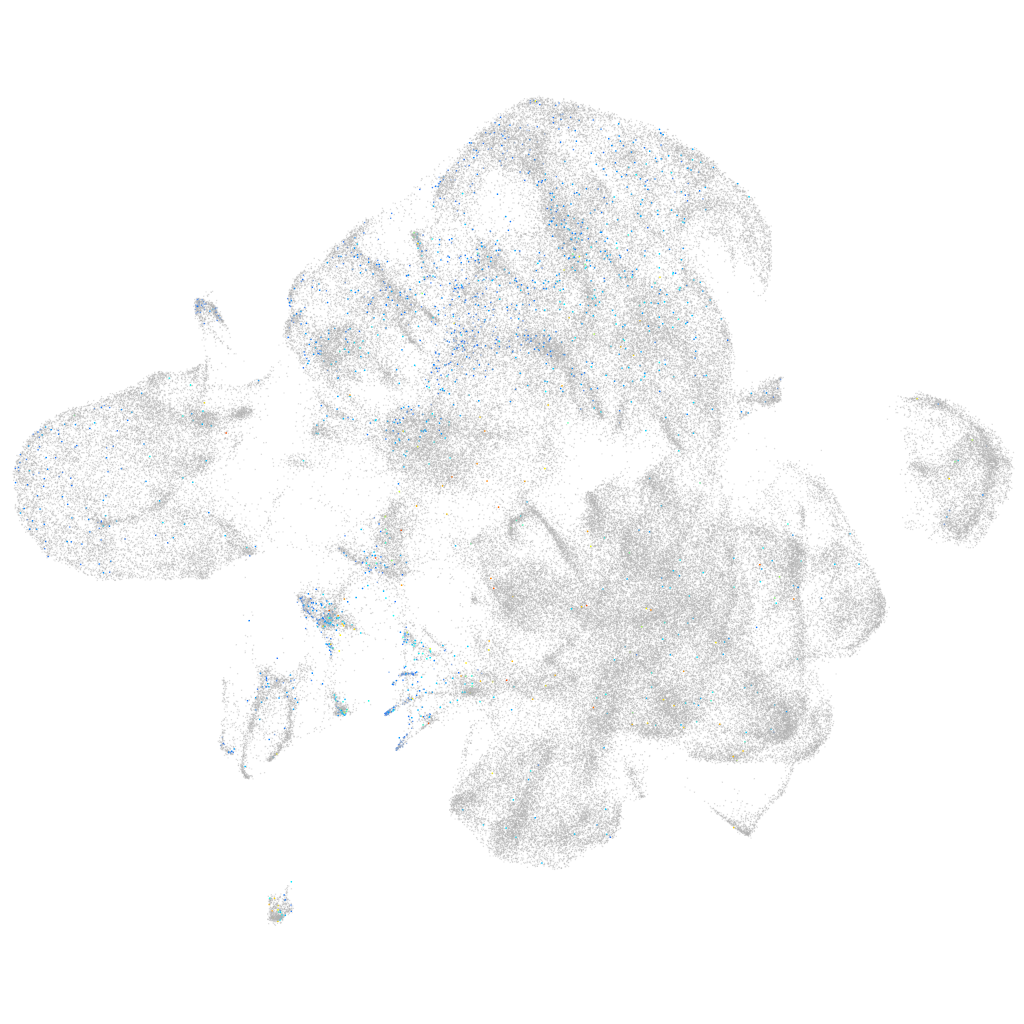
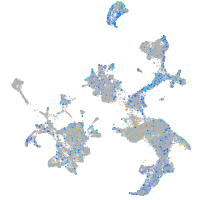
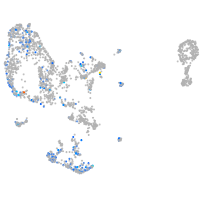
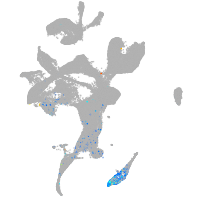
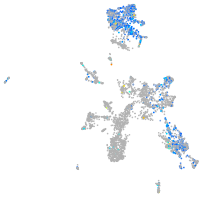
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| eef1da | 0.071 | elavl3 | -0.053 |
| fxyd1 | 0.067 | stmn1b | -0.051 |
| sparc | 0.061 | gpm6aa | -0.048 |
| crip1 | 0.058 | rtn1a | -0.047 |
| gsta.1 | 0.057 | tuba1c | -0.047 |
| sdr16c5b | 0.056 | ckbb | -0.045 |
| krt18b | 0.055 | marcksl1b | -0.045 |
| si:ch211-113d11.6 | 0.055 | zc4h2 | -0.044 |
| ahcy | 0.055 | gpm6ab | -0.044 |
| si:ch211-186e20.2 | 0.054 | tubb5 | -0.042 |
| tuba8l | 0.053 | rtn1b | -0.042 |
| anxa11a | 0.053 | sncb | -0.042 |
| eno3 | 0.053 | gng3 | -0.041 |
| anxa11b | 0.052 | ptmab | -0.040 |
| cd63 | 0.052 | hmgb3a | -0.039 |
| mhc1lia | 0.052 | nova2 | -0.039 |
| zanl | 0.052 | elavl4 | -0.038 |
| abcc2 | 0.051 | si:dkey-276j7.1 | -0.038 |
| pmp22a | 0.051 | fez1 | -0.038 |
| GCA | 0.050 | atp6v0cb | -0.037 |
| BX908782.3 | 0.050 | tmsb | -0.037 |
| apobb.1 | 0.050 | tuba1a | -0.036 |
| tm4sf4 | 0.050 | csdc2a | -0.036 |
| vil1 | 0.050 | rnasekb | -0.036 |
| lxn | 0.049 | gng2 | -0.036 |
| sdc4 | 0.049 | ptmaa | -0.035 |
| CU682777.2 | 0.049 | ywhag2 | -0.035 |
| krt8 | 0.049 | mllt11 | -0.034 |
| anxa4 | 0.049 | nsg2 | -0.034 |
| si:ch211-214p16.2 | 0.048 | stmn2a | -0.034 |
| cyp3a65 | 0.048 | stx1b | -0.034 |
| gstp1 | 0.048 | hmgb1a | -0.034 |
| apoa4b.1 | 0.048 | myt1b | -0.033 |
| lgals2b | 0.047 | id4 | -0.033 |
| stoml3b | 0.047 | dbn1 | -0.033 |