zgc:194990
ZFIN 
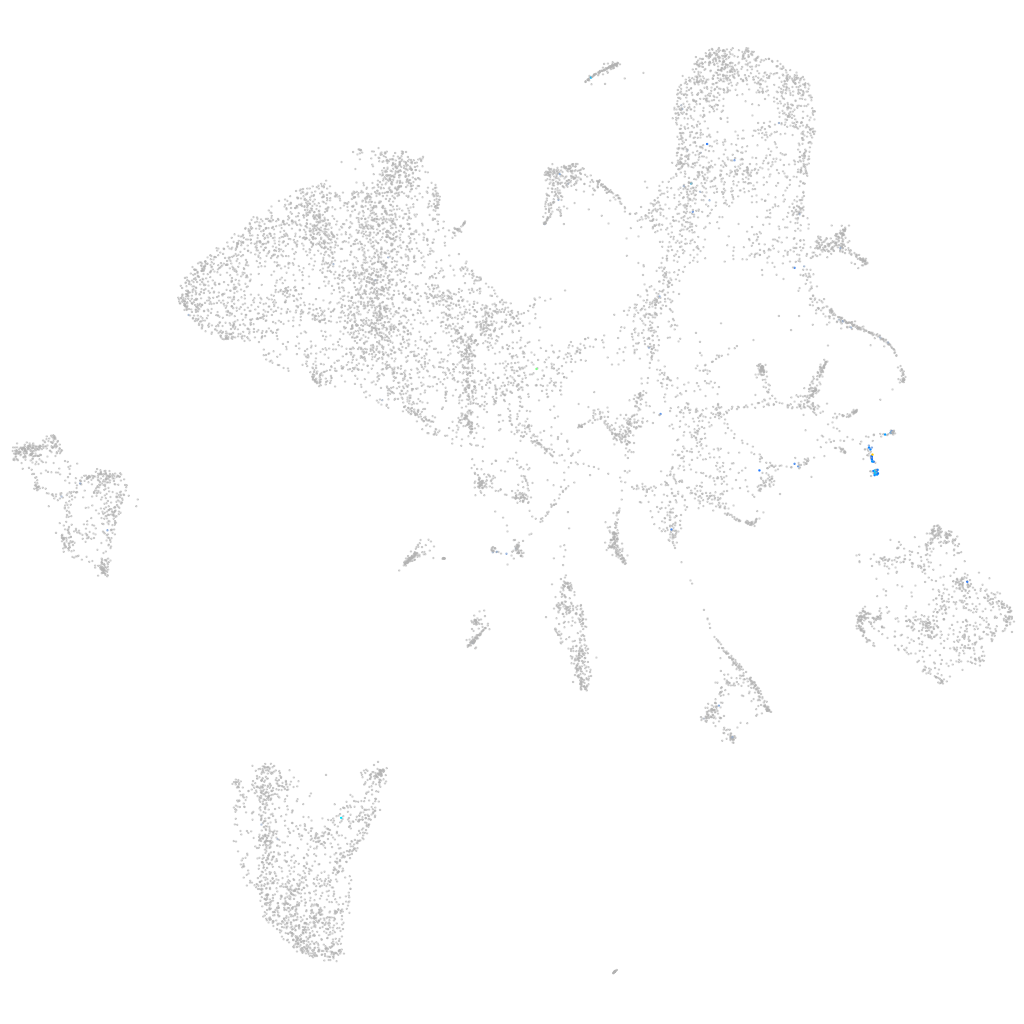
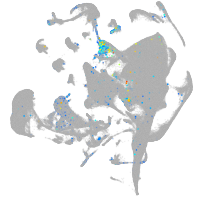
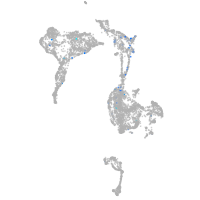
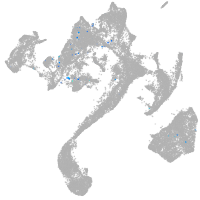
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ins | 0.427 | gapdh | -0.067 |
| ndufa4l2a | 0.425 | ahcy | -0.066 |
| slc30a2 | 0.395 | aldob | -0.062 |
| SLC5A10 | 0.394 | gamt | -0.059 |
| ucn3l | 0.381 | fbp1b | -0.057 |
| ptn | 0.377 | bhmt | -0.055 |
| rims2a | 0.312 | gpx4a | -0.054 |
| BX927333.1 | 0.297 | apoa1b | -0.053 |
| sst1.1 | 0.295 | fabp3 | -0.052 |
| gnrh2 | 0.293 | gatm | -0.052 |
| CABZ01085140.1 | 0.273 | mat1a | -0.051 |
| kcnj19a | 0.263 | glud1b | -0.051 |
| g6pcb | 0.262 | si:dkey-16p21.8 | -0.050 |
| uts1 | 0.261 | apoa4b.1 | -0.050 |
| lrfn4a | 0.260 | rps20 | -0.050 |
| cytip | 0.256 | apoc1 | -0.049 |
| pcsk2 | 0.251 | nupr1b | -0.048 |
| cpe | 0.249 | afp4 | -0.048 |
| ACVR1C | 0.247 | agxtb | -0.048 |
| pcsk1 | 0.246 | apoc2 | -0.048 |
| scgn | 0.241 | shmt1 | -0.047 |
| prokr1a | 0.236 | eno3 | -0.047 |
| AL954142.2 | 0.235 | pnp4b | -0.047 |
| scg2b | 0.232 | suclg1 | -0.047 |
| rims2b | 0.231 | aqp12 | -0.047 |
| cntnap2a | 0.224 | grhprb | -0.046 |
| CU651662.1 | 0.217 | hdlbpa | -0.046 |
| gpc1a | 0.215 | gstt1a | -0.046 |
| rrad | 0.210 | mgst1.2 | -0.046 |
| tecta | 0.209 | agxta | -0.046 |
| scg2a | 0.208 | apoa2 | -0.046 |
| tmem163a | 0.207 | aldh9a1a.1 | -0.046 |
| kcnip3b | 0.206 | cx32.3 | -0.045 |
| rtn4rl2b | 0.205 | gnmt | -0.044 |
| CABZ01085139.1 | 0.202 | tfa | -0.044 |