zgc:194281 [Source:ZFIN;Acc:ZDB-GENE-081022-94]
ZFIN 
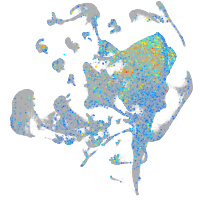
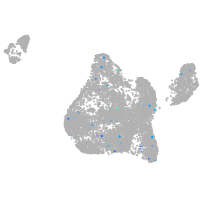
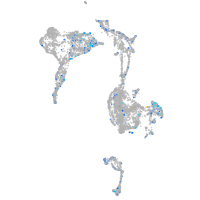
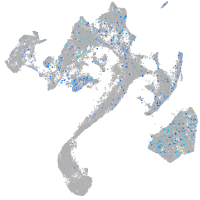
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:ch73-216m24.3 | 0.165 | gamt | -0.126 |
| wu:fj39g12 | 0.153 | gapdh | -0.122 |
| cga | 0.147 | ahcy | -0.120 |
| tshr | 0.143 | eno3 | -0.120 |
| chd7 | 0.142 | gatm | -0.112 |
| si:dkeyp-74b6.2 | 0.141 | dap | -0.109 |
| adipoqa | 0.139 | suclg1 | -0.109 |
| olfcg4 | 0.137 | fbp1b | -0.107 |
| rims2a | 0.132 | apoa4b.1 | -0.104 |
| si:dkey-27j5.5 | 0.132 | apoc2 | -0.104 |
| CABZ01066921.1 | 0.131 | mat1a | -0.103 |
| plppr4b | 0.129 | glud1b | -0.102 |
| hmgb1a | 0.127 | bhmt | -0.102 |
| aldocb | 0.126 | apoa1b | -0.102 |
| hmgn6 | 0.126 | scp2a | -0.101 |
| scg3 | 0.123 | grhprb | -0.100 |
| vat1 | 0.122 | nupr1b | -0.099 |
| marcksl1b | 0.122 | cx32.3 | -0.099 |
| si:ch211-137a8.4 | 0.122 | agxtb | -0.098 |
| LOC108191536 | 0.118 | gstt1a | -0.098 |
| neurod1 | 0.118 | aldob | -0.098 |
| si:ch211-114m9.1 | 0.117 | mgst1.2 | -0.097 |
| hmgb3a | 0.117 | sod1 | -0.097 |
| acin1a | 0.116 | pnp4b | -0.096 |
| XLOC-004668 | 0.115 | gpx4a | -0.096 |
| marcksa | 0.115 | aldh6a1 | -0.096 |
| si:ch73-1a9.3 | 0.115 | afp4 | -0.094 |
| hnrnpa0a | 0.114 | apoa2 | -0.094 |
| map1sb | 0.114 | nipsnap3a | -0.093 |
| pcsk1nl | 0.114 | agxta | -0.093 |
| mdkb | 0.114 | aqp12 | -0.092 |
| si:ch211-222l21.1 | 0.114 | COX7A2 (1 of many) | -0.092 |
| lrfn4a | 0.114 | ttc36 | -0.091 |
| syncrip | 0.113 | serpina1l | -0.091 |
| ngfrb | 0.113 | prdx2 | -0.091 |