zgc:193812
ZFIN 
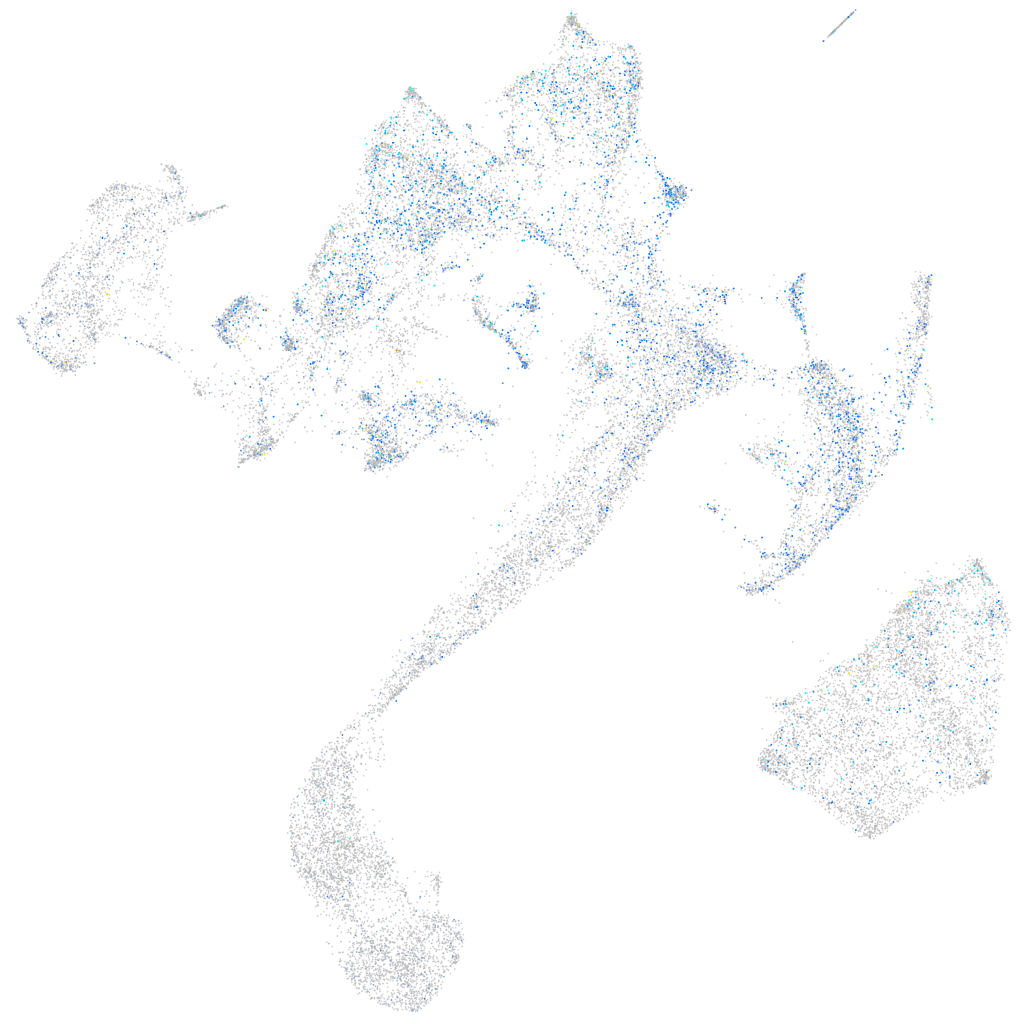
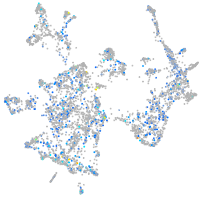
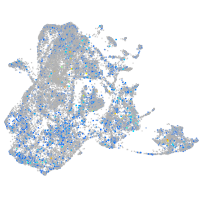
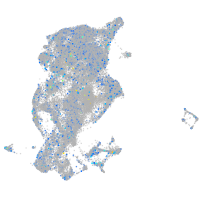
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| h3f3a | 0.142 | aldoab | -0.115 |
| marcksl1a | 0.141 | atp2a1 | -0.113 |
| mfap2 | 0.140 | tmem38a | -0.112 |
| ppiab | 0.139 | gapdh | -0.112 |
| ppib | 0.138 | tnnc2 | -0.111 |
| zgc:153867 | 0.135 | ak1 | -0.111 |
| zgc:56493 | 0.133 | acta1b | -0.111 |
| actb2 | 0.132 | ttn.1 | -0.111 |
| hnrnpa0l | 0.130 | ldb3a | -0.111 |
| pmp22a | 0.128 | ttn.2 | -0.110 |
| tubb4b | 0.127 | mybphb | -0.109 |
| dut | 0.126 | actc1b | -0.109 |
| hsp90ab1 | 0.123 | ckma | -0.108 |
| tpm4a | 0.122 | neb | -0.108 |
| dap1b | 0.121 | CABZ01078594.1 | -0.107 |
| h3f3c | 0.121 | ckmb | -0.107 |
| ptmaa | 0.119 | srl | -0.105 |
| tmsb4x | 0.117 | tpma | -0.104 |
| tmem258 | 0.117 | actn3a | -0.103 |
| ubc | 0.117 | ldb3b | -0.103 |
| nme2b.1 | 0.116 | actn3b | -0.103 |
| krt18a.1 | 0.115 | myl1 | -0.102 |
| ssr3 | 0.115 | desma | -0.100 |
| tcf15 | 0.115 | pabpc4 | -0.100 |
| zgc:92744 | 0.112 | CABZ01072309.1 | -0.100 |
| COX7A2 (1 of many) | 0.112 | txlnbb | -0.099 |
| mif | 0.112 | si:ch73-367p23.2 | -0.099 |
| rps3a | 0.112 | hhatla | -0.099 |
| actb1 | 0.111 | tnnt3a | -0.098 |
| cfl1 | 0.111 | myom1a | -0.098 |
| rpl10 | 0.109 | mylpfa | -0.098 |
| twist1a | 0.109 | smyd1a | -0.097 |
| rpl10a | 0.109 | tmod4 | -0.097 |
| rps2 | 0.108 | rtn2a | -0.097 |
| ran | 0.108 | si:ch211-266g18.10 | -0.096 |