zgc:174888
ZFIN 
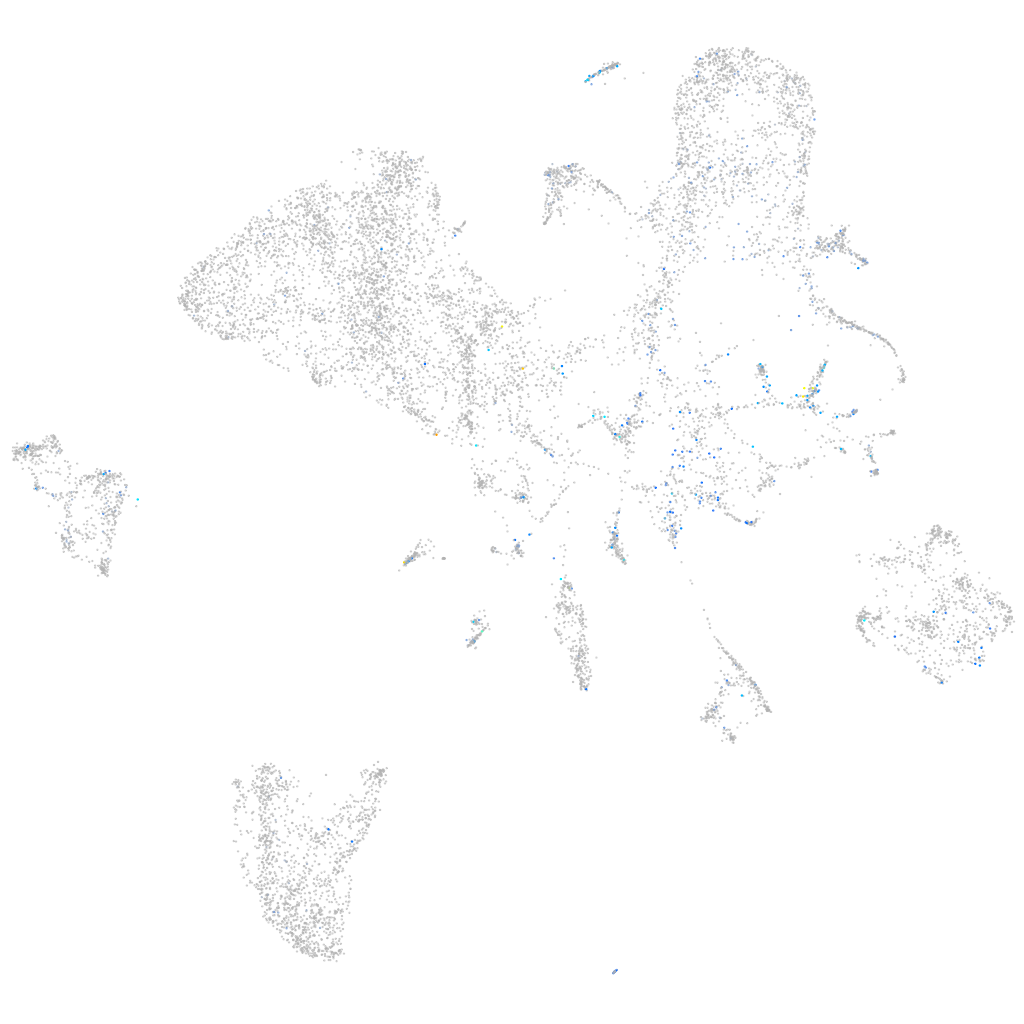
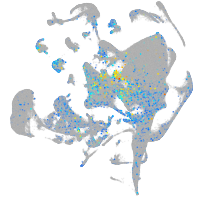
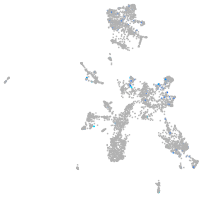
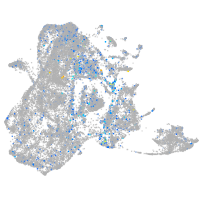
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:dkey-84h14.1 | 0.164 | gamt | -0.113 |
| XLOC-037388 | 0.155 | gatm | -0.112 |
| hepacam2 | 0.150 | mat1a | -0.109 |
| egr4 | 0.147 | bhmt | -0.109 |
| scg3 | 0.146 | gapdh | -0.104 |
| fev | 0.140 | agxtb | -0.099 |
| lysmd2 | 0.134 | apoa1b | -0.097 |
| insm1b | 0.131 | afp4 | -0.096 |
| si:ch211-216p19.5 | 0.129 | ahcy | -0.093 |
| spon1b | 0.128 | abat | -0.092 |
| zgc:165481 | 0.127 | cx32.3 | -0.092 |
| ubc | 0.123 | aqp12 | -0.091 |
| anxa4 | 0.123 | rbp2b | -0.091 |
| slc7a14a | 0.122 | nupr1b | -0.091 |
| si:dkey-108k24.2 | 0.122 | apoa2 | -0.091 |
| cib1 | 0.122 | apoc1 | -0.091 |
| rem2 | 0.122 | zgc:123103 | -0.091 |
| si:ch1073-429i10.3.1 | 0.120 | fabp10a | -0.091 |
| tspan7 | 0.119 | ttc36 | -0.090 |
| gfra3 | 0.118 | LOC110437731 | -0.090 |
| ier2b | 0.118 | apoa4b.1 | -0.089 |
| dusp2 | 0.117 | apom | -0.089 |
| rasd4 | 0.116 | tfa | -0.089 |
| mir375-2 | 0.116 | pnp4b | -0.089 |
| scgn | 0.115 | uox | -0.088 |
| tspan13b | 0.115 | ambp | -0.088 |
| capns1b | 0.115 | agxta | -0.088 |
| dok6 | 0.113 | kng1 | -0.088 |
| neurod1 | 0.112 | serpina1l | -0.088 |
| gapdhs | 0.112 | f2 | -0.088 |
| c2cd4a | 0.110 | grhprb | -0.087 |
| jun | 0.110 | fgg | -0.087 |
| ptger3 | 0.109 | fgb | -0.087 |
| jpt1b | 0.109 | ces2 | -0.087 |
| si:dkey-42i9.4 | 0.109 | ttr | -0.087 |