zgc:174263
ZFIN 
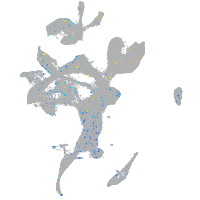
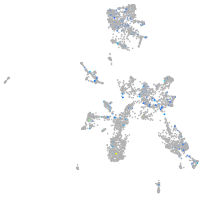
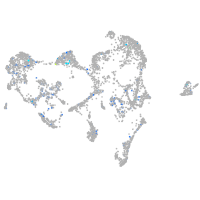
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:ch73-1a9.3 | 0.077 | actc1b | -0.068 |
| LOC100334341 | 0.077 | acta1b | -0.062 |
| seta | 0.076 | ak1 | -0.062 |
| hnrnpub | 0.076 | tnnc2 | -0.061 |
| khdrbs1a | 0.076 | tpma | -0.060 |
| nono | 0.074 | atp2a1 | -0.059 |
| hmga1a | 0.073 | ldb3a | -0.059 |
| anp32a | 0.073 | ckma | -0.059 |
| hnrnpabb | 0.073 | neb | -0.058 |
| hnrnpaba | 0.073 | ckmb | -0.058 |
| si:ch211-288g17.3 | 0.072 | mylpfa | -0.058 |
| smarca4a | 0.072 | tmem38a | -0.058 |
| si:dkey-67c22.2 | 0.072 | smyd1a | -0.058 |
| hnrnpa0b | 0.072 | aldoab | -0.058 |
| cbx3a | 0.072 | mybphb | -0.057 |
| pnrc2 | 0.072 | ttn.2 | -0.056 |
| hmgb2a | 0.072 | ldb3b | -0.055 |
| cx43.4 | 0.071 | actn3a | -0.055 |
| hmgb2b | 0.071 | cav3 | -0.055 |
| ncl | 0.071 | CABZ01078594.1 | -0.055 |
| hnrnpa1b | 0.071 | si:ch73-367p23.2 | -0.054 |
| si:ch211-222l21.1 | 0.070 | ttn.1 | -0.054 |
| syncrip | 0.070 | pvalb1 | -0.054 |
| ilf2 | 0.070 | hhatla | -0.054 |
| rbm4.3 | 0.070 | myom1a | -0.053 |
| nop56 | 0.070 | pvalb2 | -0.053 |
| si:ch73-281n10.2 | 0.069 | srl | -0.053 |
| hmgb1b | 0.069 | mylz3 | -0.053 |
| nucks1a | 0.069 | tmod4 | -0.053 |
| marcksb | 0.069 | actn3b | -0.052 |
| gnb1b | 0.069 | pgam2 | -0.052 |
| ctnnb1 | 0.069 | mylpfb | -0.052 |
| cirbpa | 0.069 | tnnt3a | -0.051 |
| si:ch211-51e12.7 | 0.069 | eno3 | -0.051 |
| ilf3b | 0.069 | nme2b.2 | -0.051 |