zgc:173705
ZFIN 
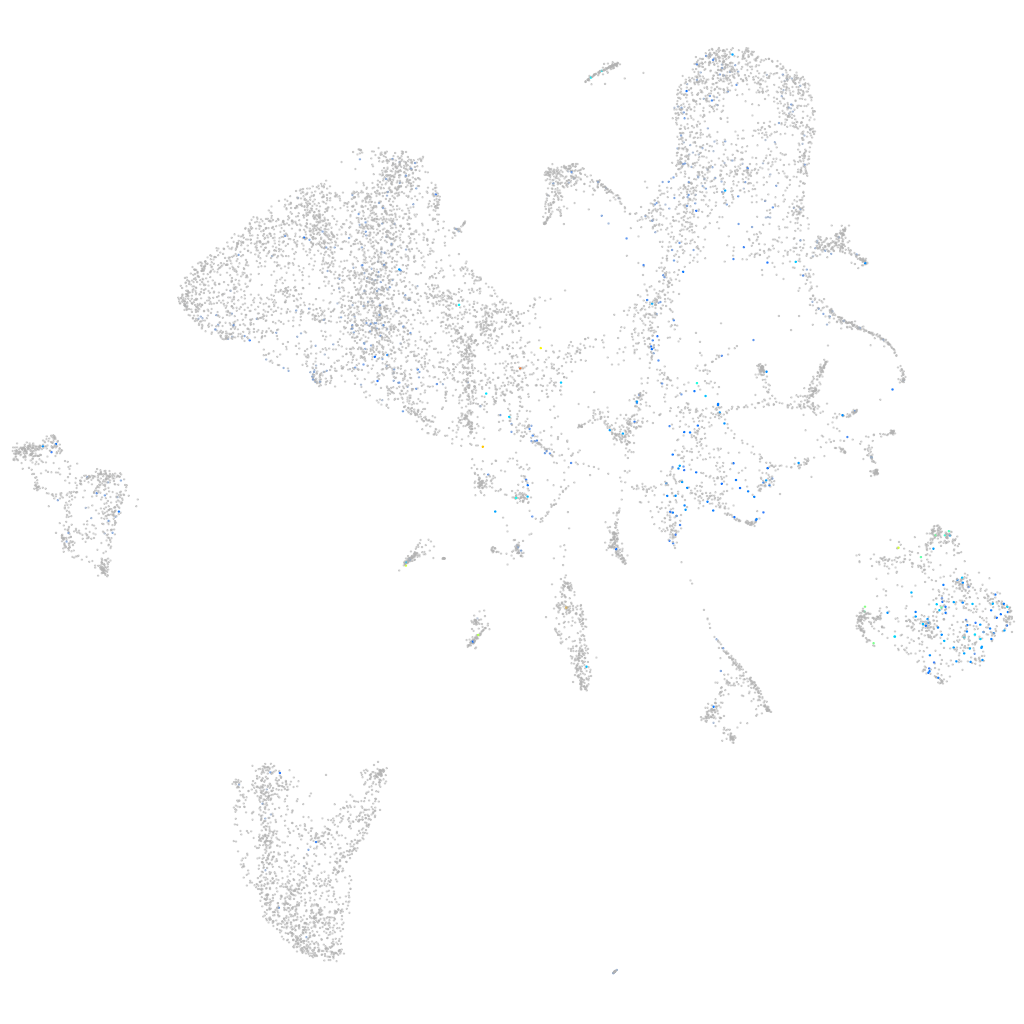
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ilf3b | 0.160 | rpl37 | -0.102 |
| ppm1g | 0.160 | rps10 | -0.096 |
| si:ch211-271b14.1 | 0.159 | zgc:114188 | -0.093 |
| hnrnpabb | 0.159 | gamt | -0.089 |
| si:ch73-281n10.2 | 0.157 | zgc:92744 | -0.089 |
| marcksl1b | 0.157 | gatm | -0.088 |
| syncrip | 0.157 | nupr1b | -0.085 |
| cx43.4 | 0.157 | rps17 | -0.084 |
| marcksb | 0.156 | bhmt | -0.084 |
| smc1al | 0.156 | zgc:158463 | -0.081 |
| hnrnpa0b | 0.156 | gapdh | -0.081 |
| hmga1a | 0.155 | nme2b.1 | -0.078 |
| khdrbs1a | 0.154 | ahcy | -0.077 |
| hnrnpub | 0.153 | dap | -0.076 |
| snrpd3l | 0.153 | pnp4b | -0.071 |
| anp32b | 0.152 | mat1a | -0.070 |
| snrnp70 | 0.152 | aqp12 | -0.069 |
| mcm6 | 0.151 | agxtb | -0.069 |
| cbx5 | 0.150 | sod1 | -0.068 |
| si:ch211-152c2.3 | 0.150 | atp5if1b | -0.068 |
| cbx3a | 0.150 | suclg1 | -0.066 |
| setb | 0.149 | eno3 | -0.066 |
| nucks1b | 0.149 | apoa1b | -0.065 |
| sae1 | 0.149 | apoa2 | -0.063 |
| ncl | 0.148 | gpx4a | -0.063 |
| seta | 0.148 | pvalb1 | -0.063 |
| nop58 | 0.148 | ckba | -0.063 |
| sp5l | 0.148 | atp5l | -0.062 |
| hdac1 | 0.148 | fbp1b | -0.062 |
| smo | 0.148 | eef1da | -0.062 |
| akap12b | 0.147 | prss1 | -0.062 |
| baz1b | 0.147 | cox7a1 | -0.062 |
| ppig | 0.147 | rbp4 | -0.061 |
| smarcb1b | 0.147 | serp1 | -0.061 |
| hdgfl2 | 0.147 | grhprb | -0.061 |