zgc:172323
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm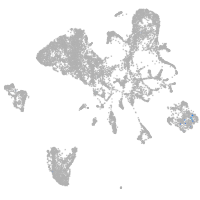 |
axial mesoderm |
muscle 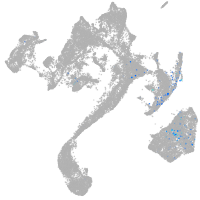 |
mural cells / non-skeletal muscle 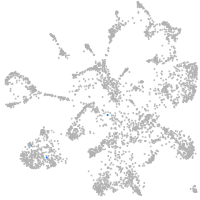 |
mesenchyme 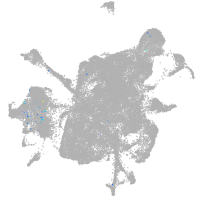 |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| prdm13 | 0.432 | hmgb2a | -0.102 |
| nhlh2 | 0.364 | msna | -0.075 |
| mir181b-3 | 0.266 | msi1 | -0.069 |
| rnd1a | 0.248 | crx | -0.068 |
| slc32a1 | 0.239 | pcna | -0.068 |
| pax6b | 0.235 | tubb4b | -0.068 |
| slit1b | 0.232 | tuba8l4 | -0.067 |
| gbx2 | 0.229 | syt5b | -0.067 |
| slc6a9 | 0.227 | tuba8l | -0.066 |
| camkvb | 0.214 | otx5 | -0.066 |
| pax6a | 0.198 | dut | -0.065 |
| si:ch73-290k24.5 | 0.184 | mki67 | -0.065 |
| phlda3 | 0.182 | eef1da | -0.064 |
| CR774179.5 | 0.169 | selenoh | -0.064 |
| syt1a | 0.162 | rrm1 | -0.063 |
| grin1a | 0.160 | stmn1a | -0.063 |
| elavl3 | 0.156 | her15.1 | -0.062 |
| shisa9a | 0.154 | banf1 | -0.061 |
| ndrg2 | 0.153 | id1 | -0.061 |
| roraa | 0.152 | ccnd1 | -0.061 |
| rbfox2 | 0.152 | ccna2 | -0.060 |
| slc30a2 | 0.150 | nutf2l | -0.059 |
| rnd1b | 0.146 | chaf1a | -0.059 |
| tcf4 | 0.146 | ahcy | -0.059 |
| si:dkey-46i9.1 | 0.145 | histh1l | -0.058 |
| zgc:165603 | 0.144 | chd7 | -0.058 |
| grin2aa | 0.143 | cks1b | -0.057 |
| ebf1a | 0.143 | mcm7 | -0.057 |
| syt9b | 0.143 | ddah2 | -0.056 |
| scrt1a | 0.142 | cldn5a | -0.055 |
| gng2 | 0.140 | vsx1 | -0.055 |
| rpp25a | 0.135 | XLOC-003690 | -0.055 |
| klf12b | 0.135 | zgc:110425 | -0.055 |
| cplx3b | 0.135 | dlgap5 | -0.055 |
| nfasca | 0.131 | dek | -0.054 |




