zgc:172145
ZFIN 
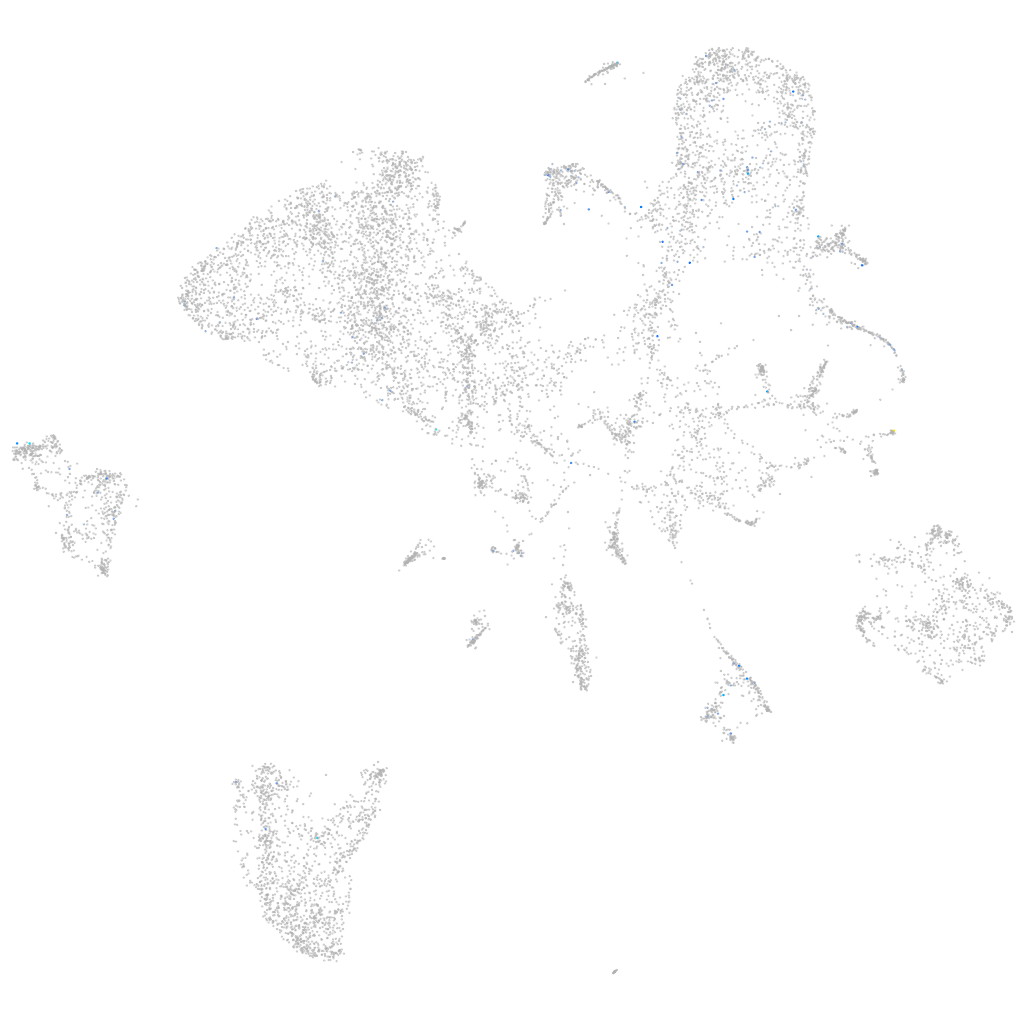
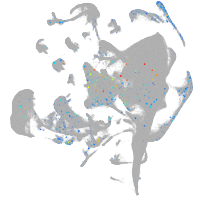
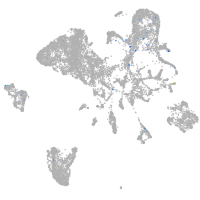
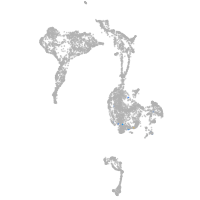
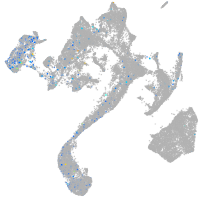
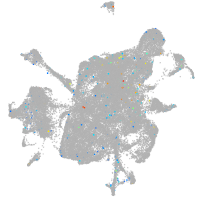
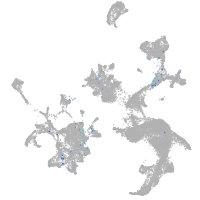
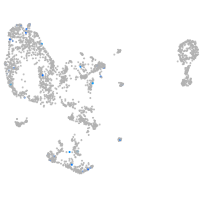
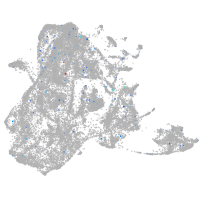
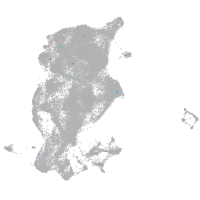
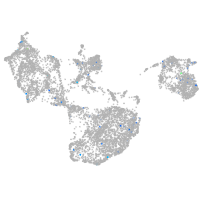
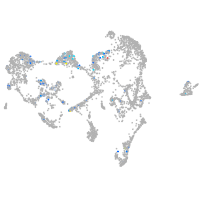
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| BX571707.1 | 0.290 | apoc1 | -0.050 |
| gpr153 | 0.231 | bhmt | -0.049 |
| LOC101885065 | 0.224 | aqp12 | -0.047 |
| pcdh2aa15 | 0.203 | gatm | -0.042 |
| mtcl1 | 0.168 | agxtb | -0.041 |
| si:ch73-233f7.6 | 0.158 | fetub | -0.040 |
| LOC108190733 | 0.154 | serpina1 | -0.040 |
| kcna6a | 0.150 | cpn1 | -0.039 |
| PITPNC1 | 0.144 | zgc:123103 | -0.039 |
| igfbp6a | 0.143 | pnp4b | -0.038 |
| si:ch211-223k15.1 | 0.136 | apoa2 | -0.038 |
| lrp2bp | 0.131 | ces2 | -0.038 |
| LOC108190355 | 0.127 | hpda | -0.038 |
| TP53I3 | 0.126 | apom | -0.037 |
| hipk1a | 0.125 | fabp3 | -0.037 |
| casr | 0.125 | fgb | -0.037 |
| pcdh1g29 | 0.124 | rbp2b | -0.037 |
| cdk5r2b | 0.121 | rbp4 | -0.037 |
| CABZ01021532.1 | 0.121 | fgg | -0.037 |
| srd5a2b | 0.119 | fabp10a | -0.037 |
| CR848025.1 | 0.116 | ambp | -0.036 |
| ccdc62 | 0.114 | gc | -0.036 |
| si:ch211-158d24.2 | 0.114 | LOC110437731 | -0.036 |
| slitrk1 | 0.112 | serpina1l | -0.036 |
| MYL2 | 0.108 | uox | -0.036 |
| ush1gb | 0.108 | cp | -0.036 |
| XLOC-020480 | 0.105 | uraha | -0.036 |
| LOC110437755 | 0.102 | grhprb | -0.036 |
| CU651657.1 | 0.101 | hao1 | -0.036 |
| CABZ01048956.1 | 0.100 | mat1a | -0.035 |
| RF01290 | 0.100 | cdab | -0.035 |
| igsf9bb | 0.099 | ttr | -0.035 |
| LOC100538221 | 0.099 | selenop2 | -0.035 |
| slc25a13 | 0.099 | vtnb | -0.035 |
| trim35-19 | 0.098 | kng1 | -0.035 |