zgc:165409
ZFIN 
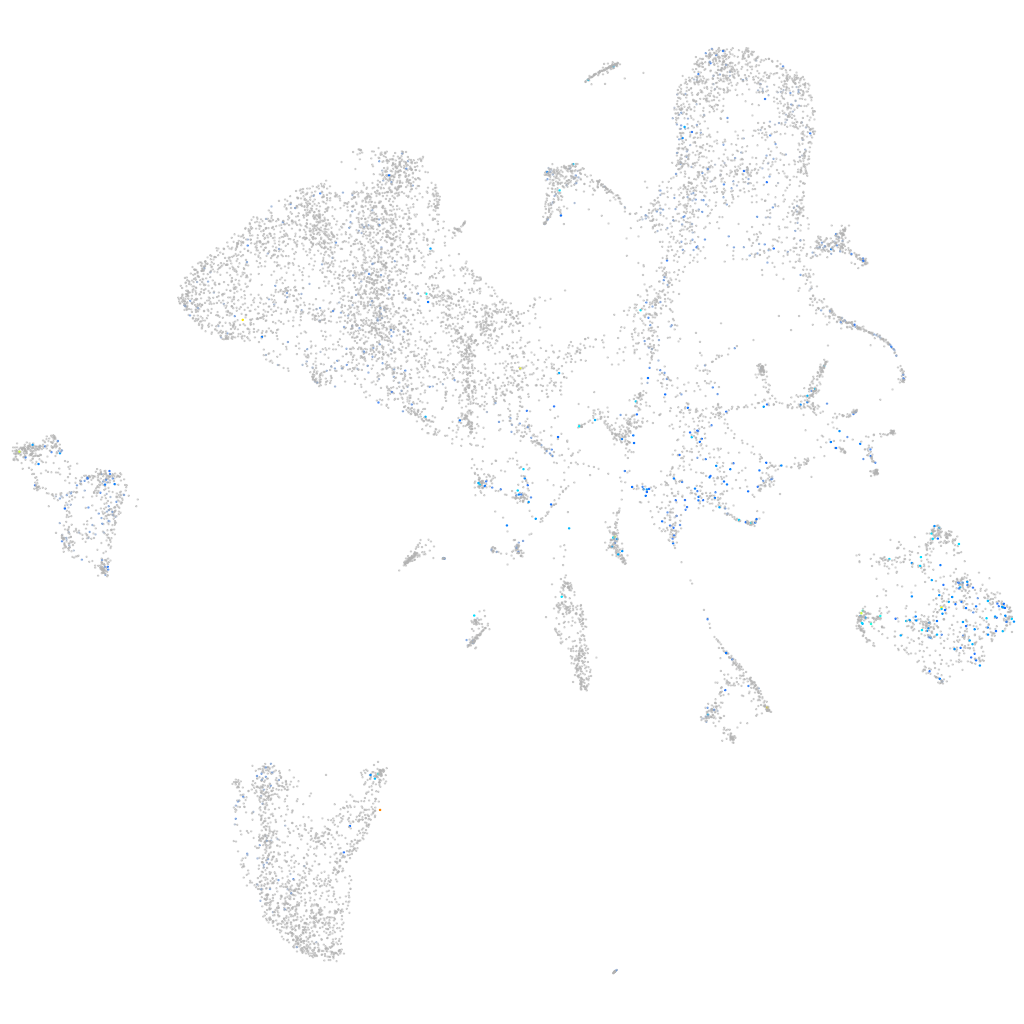
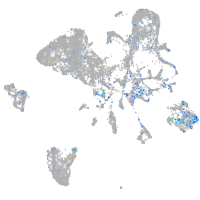
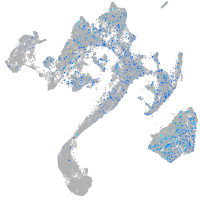
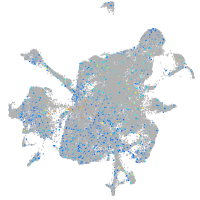
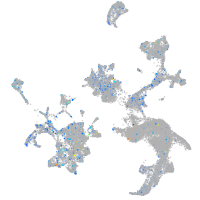
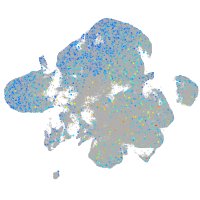
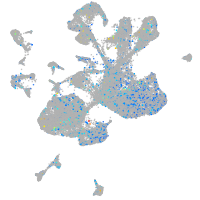
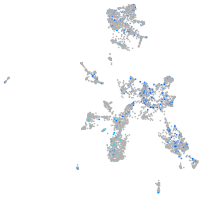
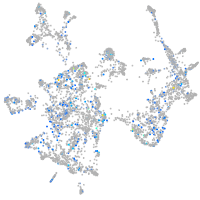
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hnrnpa0b | 0.170 | gamt | -0.125 |
| syncrip | 0.168 | gapdh | -0.122 |
| igf1 | 0.164 | gatm | -0.111 |
| hdac1 | 0.164 | bhmt | -0.110 |
| khdrbs1a | 0.161 | ahcy | -0.108 |
| hnrnpaba | 0.160 | zgc:92744 | -0.107 |
| ppig | 0.160 | pnp4b | -0.101 |
| anp32a | 0.158 | apoa1b | -0.101 |
| cbx3a | 0.157 | mat1a | -0.099 |
| hmga1a | 0.157 | fbp1b | -0.099 |
| hnrnpabb | 0.157 | rpl37 | -0.099 |
| u2af2b | 0.157 | agxtb | -0.097 |
| cx43.4 | 0.156 | apoa2 | -0.097 |
| ilf3b | 0.156 | eno3 | -0.096 |
| si:ch73-1a9.3 | 0.155 | suclg1 | -0.094 |
| marcksb | 0.155 | nupr1b | -0.094 |
| srsf3b | 0.155 | apoa4b.1 | -0.094 |
| seta | 0.155 | abat | -0.093 |
| marcksl1b | 0.155 | scp2a | -0.093 |
| nono | 0.155 | glud1b | -0.092 |
| nucks1a | 0.153 | gpx4a | -0.092 |
| hmgb2a | 0.153 | dap | -0.092 |
| si:ch73-281n10.2 | 0.153 | grhprb | -0.092 |
| ilf2 | 0.153 | aldh6a1 | -0.091 |
| LOC110438300 | 0.153 | cx32.3 | -0.091 |
| hnrnpub | 0.152 | fetub | -0.090 |
| srsf11 | 0.152 | afp4 | -0.090 |
| smc1al | 0.152 | fabp10a | -0.089 |
| srrm2 | 0.151 | ces2 | -0.088 |
| nop58 | 0.151 | apom | -0.088 |
| si:ch211-222l21.1 | 0.151 | zgc:123103 | -0.088 |
| cpsf6 | 0.151 | rps10 | -0.088 |
| acin1a | 0.150 | apoc2 | -0.087 |
| h2afvb | 0.149 | rbp2b | -0.087 |
| smarca4a | 0.149 | serpina1l | -0.086 |