zgc:162144
ZFIN 
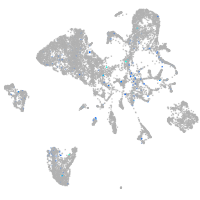
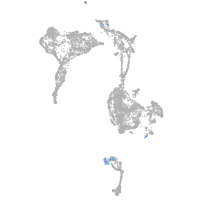
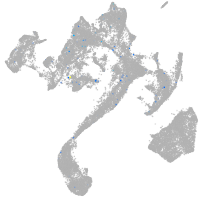
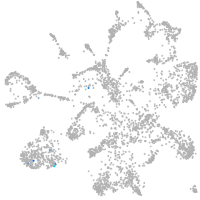
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| PLEKHB1 | 0.206 | NC-002333.4 | -0.026 |
| guca1b | 0.172 | hspb1 | -0.024 |
| saga | 0.164 | si:ch211-152c2.3 | -0.023 |
| rom1a | 0.151 | ved | -0.023 |
| cnga1b | 0.137 | hnrnpd | -0.022 |
| otol1a | 0.133 | tnnt3a | -0.022 |
| gnat1 | 0.130 | ldb3b | -0.022 |
| si:ch211-113d22.2 | 0.129 | cdx4 | -0.021 |
| rom1b | 0.126 | rbm38 | -0.021 |
| LOC103909738 | 0.121 | aldoab | -0.021 |
| nrl | 0.120 | tbx16 | -0.021 |
| sagb | 0.120 | myom1a | -0.021 |
| ccl39.2 | 0.118 | acta1b | -0.021 |
| taar20t | 0.116 | ckma | -0.020 |
| cnr2 | 0.106 | tnnc2 | -0.020 |
| cabp4 | 0.106 | ldb3a | -0.020 |
| ptprt | 0.105 | neb | -0.020 |
| pde6ga | 0.105 | mybphb | -0.020 |
| nr2e3 | 0.103 | si:ch211-266g18.10 | -0.020 |
| trim35-33 | 0.101 | dhrs7cb | -0.020 |
| rho | 0.099 | cdx1a | -0.019 |
| gc2 | 0.097 | CABZ01078594.1 | -0.019 |
| pdca | 0.097 | actn3a | -0.019 |
| gngt1 | 0.095 | COX7A2 | -0.019 |
| kcnv2a | 0.092 | XLOC-006515 | -0.019 |
| cngb1a | 0.092 | CR855257.1 | -0.019 |
| LOC108180196 | 0.090 | jph1b | -0.019 |
| XLOC-010030 | 0.088 | trdn | -0.019 |
| AL928685.3 | 0.087 | im:7138239 | -0.019 |
| kifl | 0.085 | mylz3 | -0.019 |
| pde6ha | 0.081 | ank1a | -0.019 |
| ap1m3 | 0.080 | cfl2 | -0.019 |
| slc39a4 | 0.075 | smyd1a | -0.019 |
| AL732515.1 | 0.075 | tmod4 | -0.019 |
| si:dkey-114l24.2 | 0.073 | myom2a | -0.019 |