zgc:158803
ZFIN 
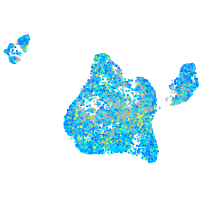
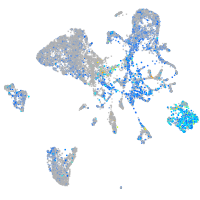
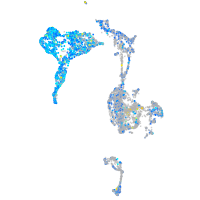
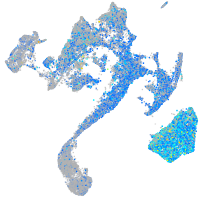
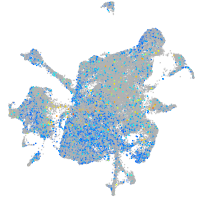
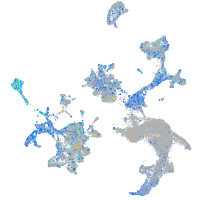
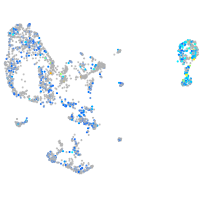
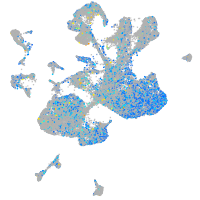
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ncl | 0.459 | rpl37 | -0.399 |
| hnrnpa1b | 0.445 | zgc:114188 | -0.371 |
| hnrnpub | 0.441 | rps10 | -0.369 |
| anp32e | 0.437 | rps17 | -0.350 |
| nop58 | 0.435 | actc1b | -0.321 |
| fbl | 0.434 | ckma | -0.291 |
| ppig | 0.427 | ak1 | -0.289 |
| dkc1 | 0.427 | vdac3 | -0.288 |
| nop56 | 0.422 | ckmb | -0.286 |
| hnrnpa1a | 0.418 | tnnc2 | -0.275 |
| rbm4.3 | 0.416 | atp5meb | -0.275 |
| npm1a | 0.416 | atp2a1 | -0.274 |
| top1l | 0.416 | tpi1b | -0.268 |
| snrnp70 | 0.415 | bhmt | -0.268 |
| nucks1a | 0.413 | mylpfa | -0.268 |
| marcksb | 0.413 | aldoab | -0.267 |
| seta | 0.412 | pvalb1 | -0.267 |
| hnrnpm | 0.412 | eno3 | -0.267 |
| ilf3b | 0.411 | mt-nd1 | -0.266 |
| acin1a | 0.410 | neb | -0.266 |
| NC-002333.4 | 0.407 | fabp3 | -0.265 |
| srrm1 | 0.406 | idh2 | -0.265 |
| syncrip | 0.405 | eef1da | -0.264 |
| srsf1a | 0.403 | acta1b | -0.262 |
| hnrnpabb | 0.403 | pvalb2 | -0.261 |
| nop2 | 0.403 | atp5f1b | -0.259 |
| bms1 | 0.402 | tpma | -0.258 |
| hmga1a | 0.400 | nme2b.2 | -0.257 |
| stm | 0.397 | ttn.2 | -0.257 |
| cdx4 | 0.395 | si:ch73-367p23.2 | -0.257 |
| brd3a | 0.395 | actn3a | -0.256 |
| safb | 0.392 | ldb3b | -0.255 |
| cirbpa | 0.392 | actn3b | -0.253 |
| cx43.4 | 0.391 | vdac2 | -0.253 |
| zmp:0000000624 | 0.389 | mylz3 | -0.253 |