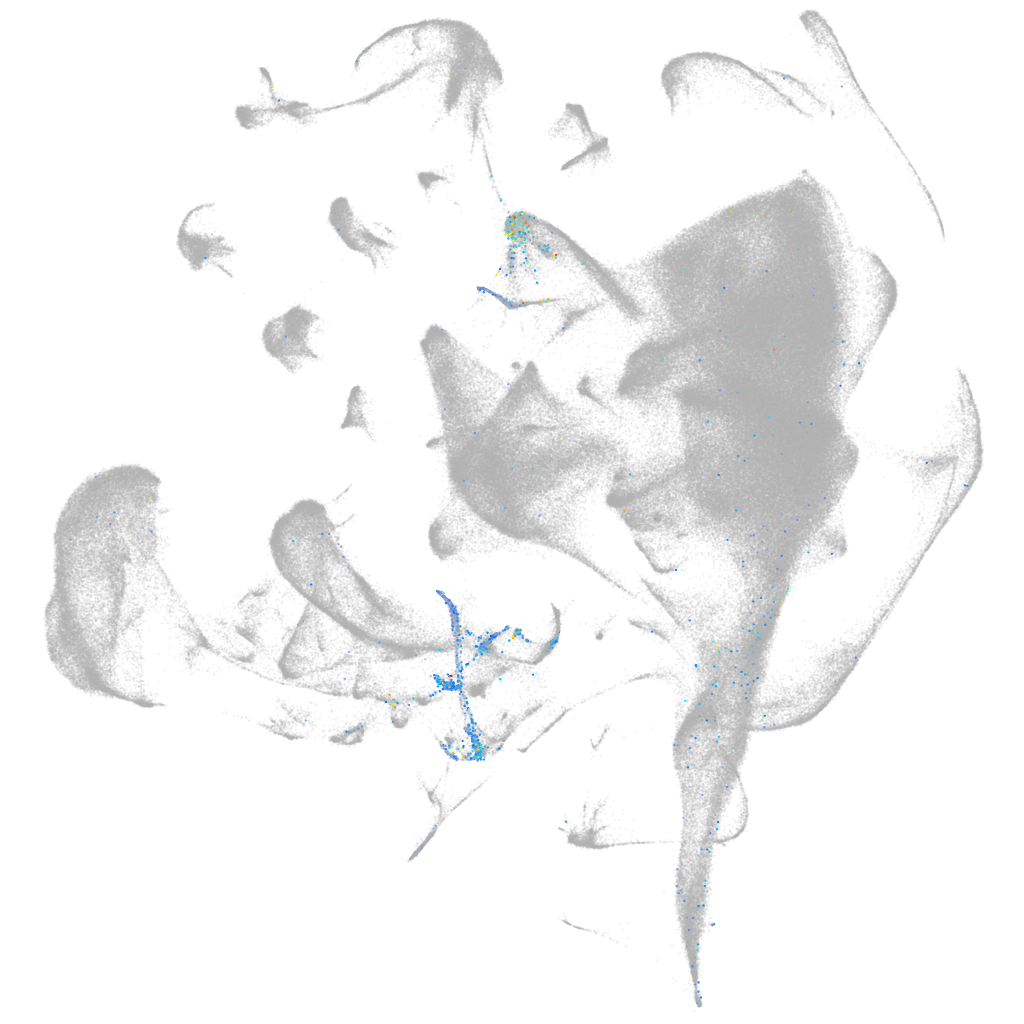
zgc:158258
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm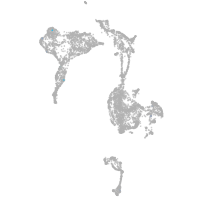 |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros 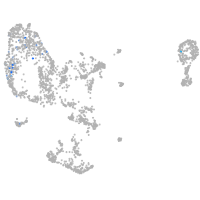 |
neural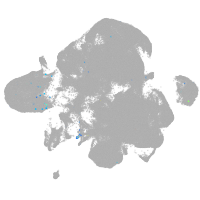 |
spinal cord / glia |
eye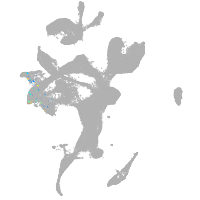 |
taste / olfactory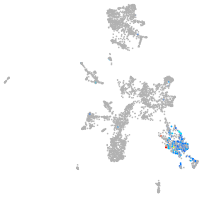 |
otic / lateral line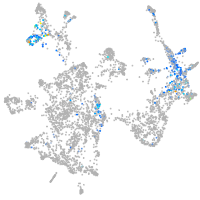 |
epidermis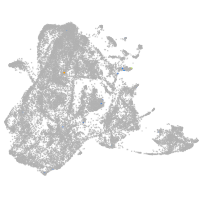 |
periderm |
fin |
ionocytes / mucous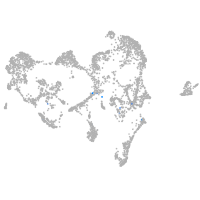 |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| calml4a | 0.243 | marcksl1a | -0.048 |
| tmem176l.2 | 0.238 | gpm6aa | -0.044 |
| moxd1l | 0.227 | gpm6ab | -0.042 |
| krt1-c5 | 0.209 | nova2 | -0.042 |
| cldn15la | 0.208 | tubb5 | -0.038 |
| plac8.1 | 0.203 | celf2 | -0.037 |
| tm4sf4 | 0.199 | mdkb | -0.037 |
| zgc:172079 | 0.199 | cspg5a | -0.035 |
| eps8l3b | 0.197 | hmgb3a | -0.034 |
| si:dkeyp-73b11.8 | 0.197 | si:dkey-56m19.5 | -0.034 |
| wu:fb59d01 | 0.195 | hmgb1b | -0.033 |
| AL831745.1 | 0.192 | tmeff1b | -0.033 |
| vil1 | 0.192 | tuba1c | -0.033 |
| anxa2b | 0.190 | elavl3 | -0.032 |
| si:ch211-133l5.7 | 0.190 | fam168a | -0.031 |
| zgc:77748 | 0.190 | mdka | -0.031 |
| anpepa | 0.187 | zc4h2 | -0.031 |
| gpa33b | 0.185 | rtn1b | -0.030 |
| ptprh | 0.184 | stmn1b | -0.030 |
| zanl | 0.183 | si:ch73-46j18.5 | -0.029 |
| LOC100331417 | 0.182 | ywhag2 | -0.029 |
| LOC103908636 | 0.182 | cd99l2 | -0.028 |
| tmc5 | 0.182 | elavl4 | -0.028 |
| clrn3 | 0.181 | cadm3 | -0.027 |
| lgals2b | 0.181 | fabp3 | -0.027 |
| si:ch211-137i24.10 | 0.181 | fscn1a | -0.027 |
| zgc:136872 | 0.180 | hbbe1.3 | -0.027 |
| adgrg4a | 0.179 | hnrnpa0a | -0.027 |
| stoml3b | 0.179 | marcksb | -0.027 |
| tspan13a | 0.179 | nat8l | -0.027 |
| cdx1b | 0.177 | ndrg4 | -0.027 |
| myo15b | 0.177 | stx1b | -0.027 |
| s100a10a | 0.176 | syt2a | -0.027 |
| adgrg4b | 0.170 | tmem59l | -0.027 |
| eps8l3a | 0.170 | dbn1 | -0.026 |