zgc:154058
ZFIN 
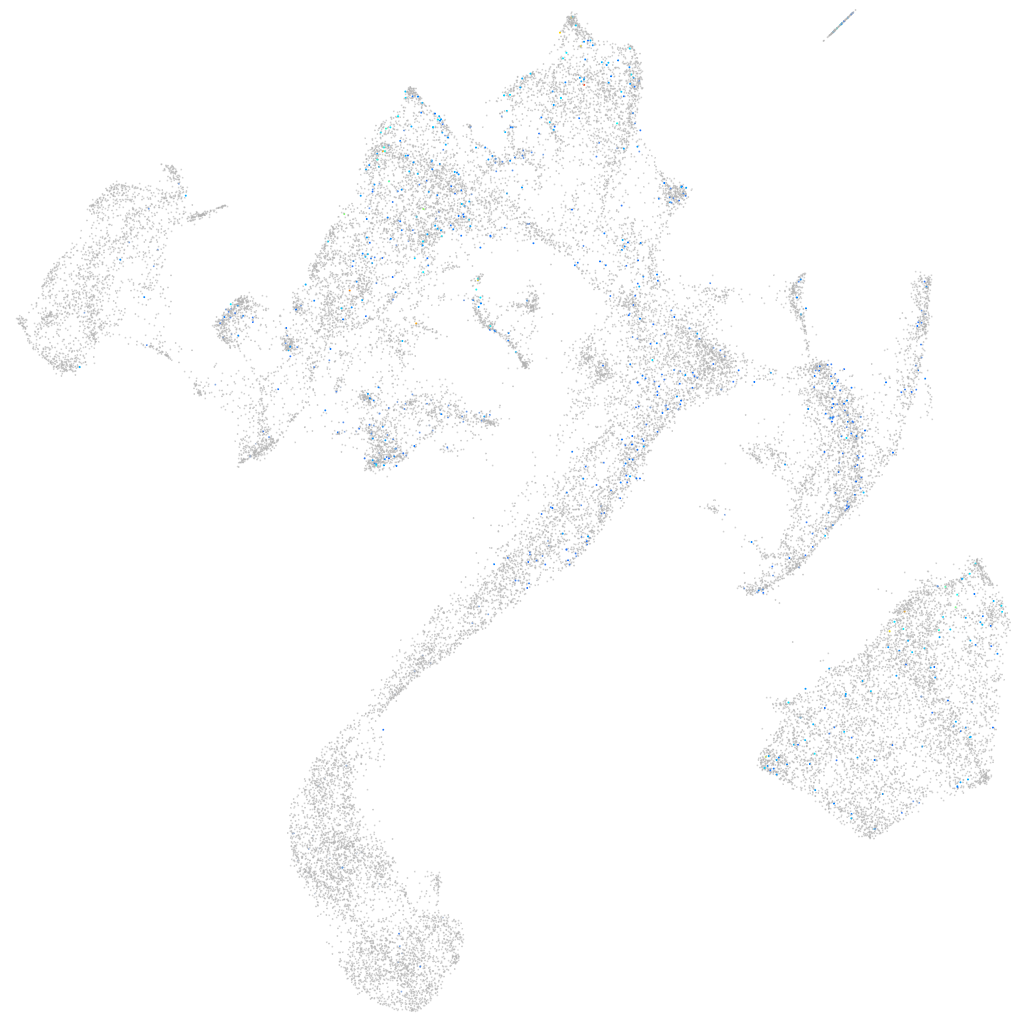
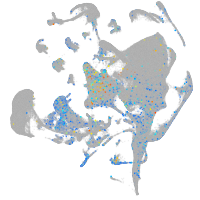
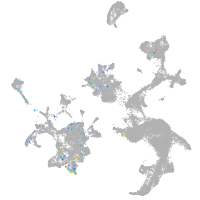
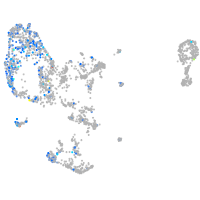
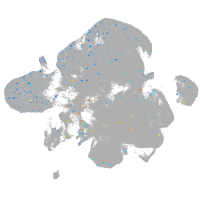
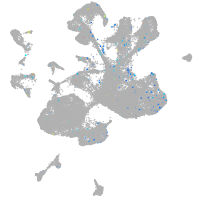
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cntn3a.2 | 0.075 | ak1 | -0.068 |
| erich3 | 0.075 | ckmb | -0.067 |
| tmsb4x | 0.073 | atp2a1 | -0.066 |
| hspb12 | 0.069 | ckma | -0.065 |
| col1a1b | 0.068 | aldoab | -0.064 |
| ppib | 0.068 | tnnc2 | -0.064 |
| XLOC-026835 | 0.067 | tmem38a | -0.063 |
| es1 | 0.066 | neb | -0.063 |
| zgc:162730 | 0.065 | gapdh | -0.062 |
| pdgfrl | 0.064 | actc1b | -0.061 |
| CR925755.2 | 0.064 | smyd1a | -0.061 |
| postnb | 0.064 | tpma | -0.061 |
| hsp90ab1 | 0.063 | si:ch73-367p23.2 | -0.061 |
| mfap2 | 0.063 | actn3b | -0.060 |
| col5a1 | 0.063 | tnnt3a | -0.060 |
| BX511034.2 | 0.063 | tmod4 | -0.059 |
| sparc | 0.062 | ldb3b | -0.059 |
| col1a2 | 0.062 | actn3a | -0.059 |
| clca1 | 0.062 | myom1a | -0.059 |
| epn3b | 0.061 | ank1a | -0.059 |
| zgc:153867 | 0.061 | cav3 | -0.059 |
| marcksl1a | 0.061 | pgam2 | -0.058 |
| BX296526.2 | 0.061 | hhatla | -0.058 |
| actb2 | 0.060 | CABZ01078594.1 | -0.056 |
| slc8a1a | 0.060 | srl | -0.056 |
| gpm6ab | 0.060 | casq2 | -0.055 |
| kcnj1a.6 | 0.060 | mylpfa | -0.055 |
| col5a2a | 0.060 | cavin4b | -0.055 |
| XLOC-043760 | 0.060 | eno3 | -0.055 |
| mxra8b | 0.059 | acta1b | -0.055 |
| tpm4a | 0.059 | mybphb | -0.055 |
| si:ch1073-459j12.1 | 0.059 | casq1b | -0.055 |
| col1a1a | 0.058 | tmem182a | -0.054 |
| foxp4 | 0.058 | pabpc4 | -0.054 |
| krt8 | 0.058 | nme2b.2 | -0.054 |