zgc:123103
ZFIN 
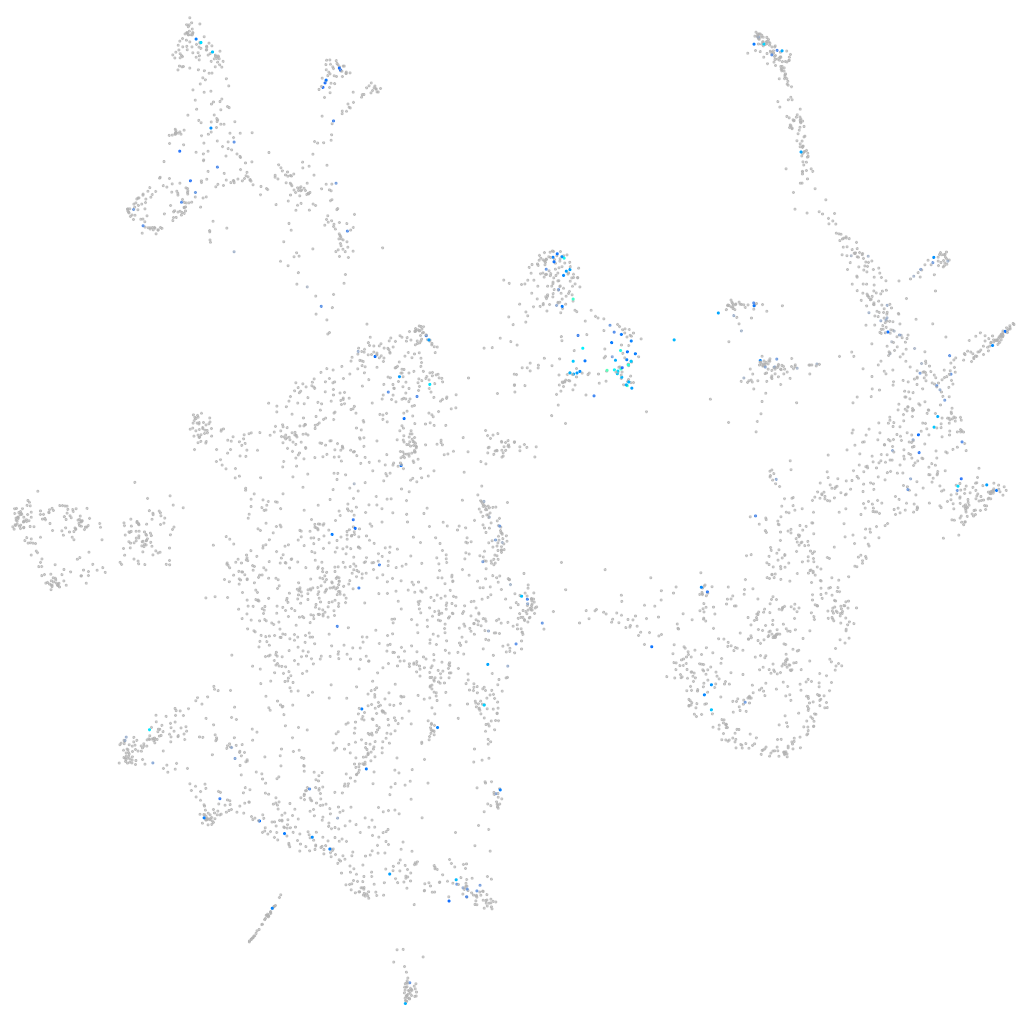
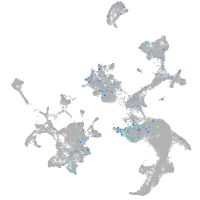
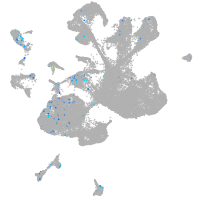
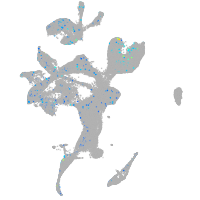
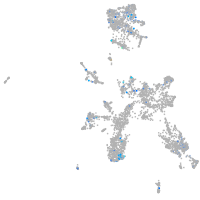
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| apoa1b | 0.410 | hsp90ab1 | -0.119 |
| apoa2 | 0.388 | serbp1a | -0.118 |
| tfa | 0.343 | eif3ea | -0.106 |
| apom | 0.338 | snrpb | -0.106 |
| fabp10a | 0.314 | zgc:153409 | -0.104 |
| apoa4b.1 | 0.311 | si:ch211-222l21.1 | -0.100 |
| serpina1l | 0.295 | six1b | -0.100 |
| krt17 | 0.284 | rpl3 | -0.097 |
| rbp4 | 0.275 | igf2bp1 | -0.095 |
| prss59.2 | 0.274 | nucks1a | -0.094 |
| lyz | 0.272 | fbl | -0.092 |
| serpina1 | 0.259 | rps11 | -0.092 |
| fetub | 0.255 | id1 | -0.092 |
| icn2 | 0.252 | snrpg | -0.091 |
| lgals2b | 0.252 | acin1a | -0.091 |
| ces2 | 0.250 | rps18 | -0.091 |
| si:ch211-270n8.1 | 0.242 | seta | -0.091 |
| fgb | 0.242 | apex1 | -0.090 |
| prss1 | 0.242 | hmga1a | -0.090 |
| gapdh | 0.234 | psmd8 | -0.088 |
| serpinf2b | 0.227 | ptges3b | -0.088 |
| apoc1 | 0.222 | sub1a | -0.087 |
| tcnba | 0.222 | pabpc1a | -0.087 |
| rbp2b | 0.222 | rpl13 | -0.087 |
| pvalb1 | 0.218 | rpl23a | -0.087 |
| afp4 | 0.216 | abcf1 | -0.087 |
| cfhl4 | 0.214 | eny2 | -0.086 |
| actc1b | 0.214 | snrpd1 | -0.084 |
| hbbe1.1 | 0.209 | hnrnpa1b | -0.084 |
| gng13a | 0.207 | sae1 | -0.083 |
| pvalb2 | 0.204 | six1a | -0.082 |
| g6pca.2 | 0.202 | psma4 | -0.082 |
| ambp | 0.202 | eya1 | -0.082 |
| prss59.1 | 0.198 | eif3ha | -0.082 |
| hbbe2 | 0.198 | ammecr1 | -0.081 |