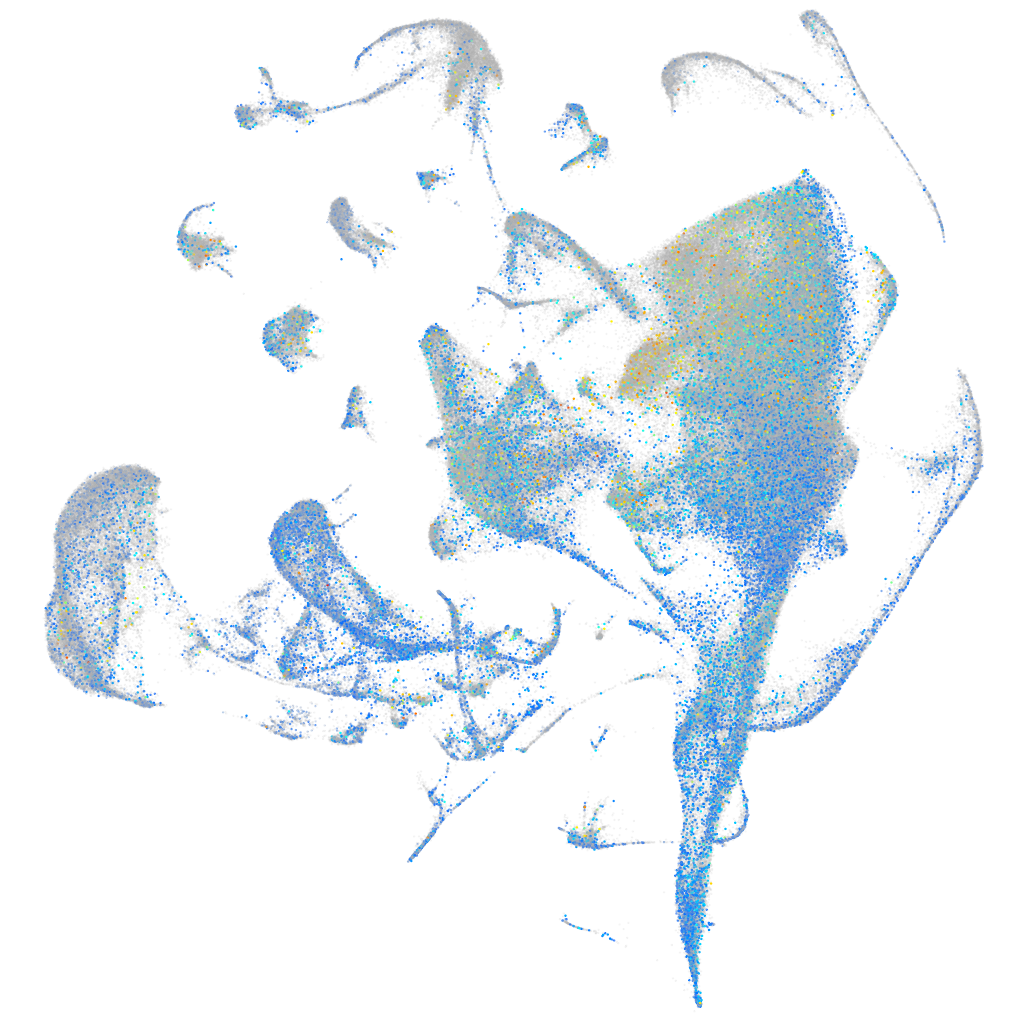zgc:114200
ZFIN 
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| id1 | 0.054 | actc1b | -0.040 |
| wls | 0.053 | pvalb1 | -0.039 |
| apoeb | 0.052 | pvalb2 | -0.037 |
| bzw1b | 0.052 | hbae3 | -0.035 |
| cx43.4 | 0.052 | hbbe1.1 | -0.035 |
| akap12b | 0.051 | hbbe2 | -0.035 |
| ctnnb1 | 0.049 | hbae1.1 | -0.034 |
| sdc4 | 0.049 | hbae1.3 | -0.033 |
| tspan7 | 0.049 | hbbe1.3 | -0.033 |
| gng5 | 0.048 | hbbe1.2 | -0.031 |
| cdc42l | 0.047 | alas2 | -0.030 |
| col18a1a | 0.047 | mylpfa | -0.030 |
| eif5a2 | 0.047 | tmod4 | -0.030 |
| ptbp1b | 0.047 | tpma | -0.030 |
| ctsla | 0.046 | cahz | -0.029 |
| snrpb | 0.046 | mylz3 | -0.029 |
| srsf5a | 0.046 | syt5b | -0.029 |
| acin1a | 0.045 | ckma | -0.028 |
| hnrnpa1b | 0.045 | hemgn | -0.028 |
| nucks1a | 0.045 | slc4a1a | -0.028 |
| smarca4a | 0.045 | zgc:163057 | -0.028 |
| chd7 | 0.044 | icn2 | -0.027 |
| igf2bp1 | 0.044 | krt17 | -0.027 |
| smc1al | 0.044 | mylpfb | -0.027 |
| cd151l | 0.043 | nt5c2l1 | -0.027 |
| etf1b | 0.043 | plac8l1 | -0.027 |
| glulb | 0.043 | si:ch211-207c6.2 | -0.027 |
| rbm4.3 | 0.043 | si:ch211-250g4.3 | -0.027 |
| rras | 0.043 | tcnbb | -0.027 |
| taf15 | 0.043 | tnnc2 | -0.027 |
| cbx1a | 0.042 | atp2a1 | -0.027 |
| fermt1 | 0.042 | agr1 | -0.026 |
| hdac1 | 0.042 | ckmb | -0.026 |
| hnrnpub | 0.042 | epb41b | -0.026 |
| kctd5a | 0.042 | hrc | -0.026 |