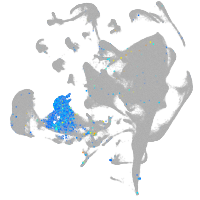
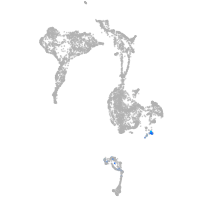
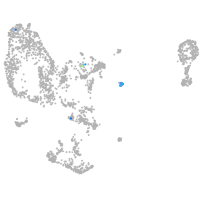
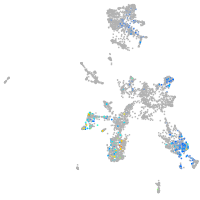
zgc:113223
ZFIN 
Other cell groups


Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| mir375-2 | 0.436 | eef1a1l1 | -0.067 |
| AL929435.1 | 0.419 | rpl12 | -0.047 |
| tagapa | 0.413 | rps26l | -0.047 |
| shisa3 | 0.392 | uba52 | -0.043 |
| fermt1 | 0.371 | rps27a | -0.042 |
| ptprfa | 0.359 | rpl13 | -0.040 |
| thbs1a | 0.309 | rps11 | -0.039 |
| or137-8 | 0.305 | actb2 | -0.039 |
| tle2b | 0.296 | rps15 | -0.038 |
| ddb2 | 0.285 | rps5 | -0.036 |
| LOC101883052 | 0.284 | rpl36 | -0.034 |
| XLOC-000431 | 0.277 | slc25a5 | -0.034 |
| tp63 | 0.271 | rpl9 | -0.032 |
| XLOC-021253 | 0.270 | zgc:114188 | -0.030 |
| XLOC-042485 | 0.270 | mt-co1 | -0.027 |
| CABZ01090219.1 | 0.263 | rpl15 | -0.027 |
| edaradd | 0.256 | rps6 | -0.025 |
| alcamb | 0.227 | qdpra | -0.024 |
| si:ch211-210b2.2 | 0.224 | rpl3 | -0.023 |
| LOC103910731 | 0.222 | tpt1 | -0.023 |
| rln3a | 0.221 | marcksl1b | -0.023 |
| vars2 | 0.221 | zgc:56493 | -0.023 |
| nudt15 | 0.207 | cfl1 | -0.022 |
| rrbp1b | 0.206 | eef2b | -0.022 |
| cd99 | 0.205 | fabp3 | -0.022 |
| CT573103.1 | 0.200 | LOC103910009 | -0.020 |
| LO018629.1 | 0.199 | syngr1a | -0.020 |
| si:dkey-204l11.1 | 0.199 | eif3f | -0.020 |
| XLOC-040020 | 0.198 | tspan10 | -0.020 |
| XLOC-034521 | 0.196 | rabl6b | -0.020 |
| CU570682.1 | 0.194 | atp6ap2 | -0.019 |
| rln3b | 0.189 | mt-cyb | -0.019 |
| col4a6 | 0.187 | gstp1 | -0.019 |
| si:dkeyp-69c1.6 | 0.182 | pcdh10a | -0.019 |
| trmt2a | 0.178 | myo5aa | -0.019 |