zgc:110591
ZFIN 
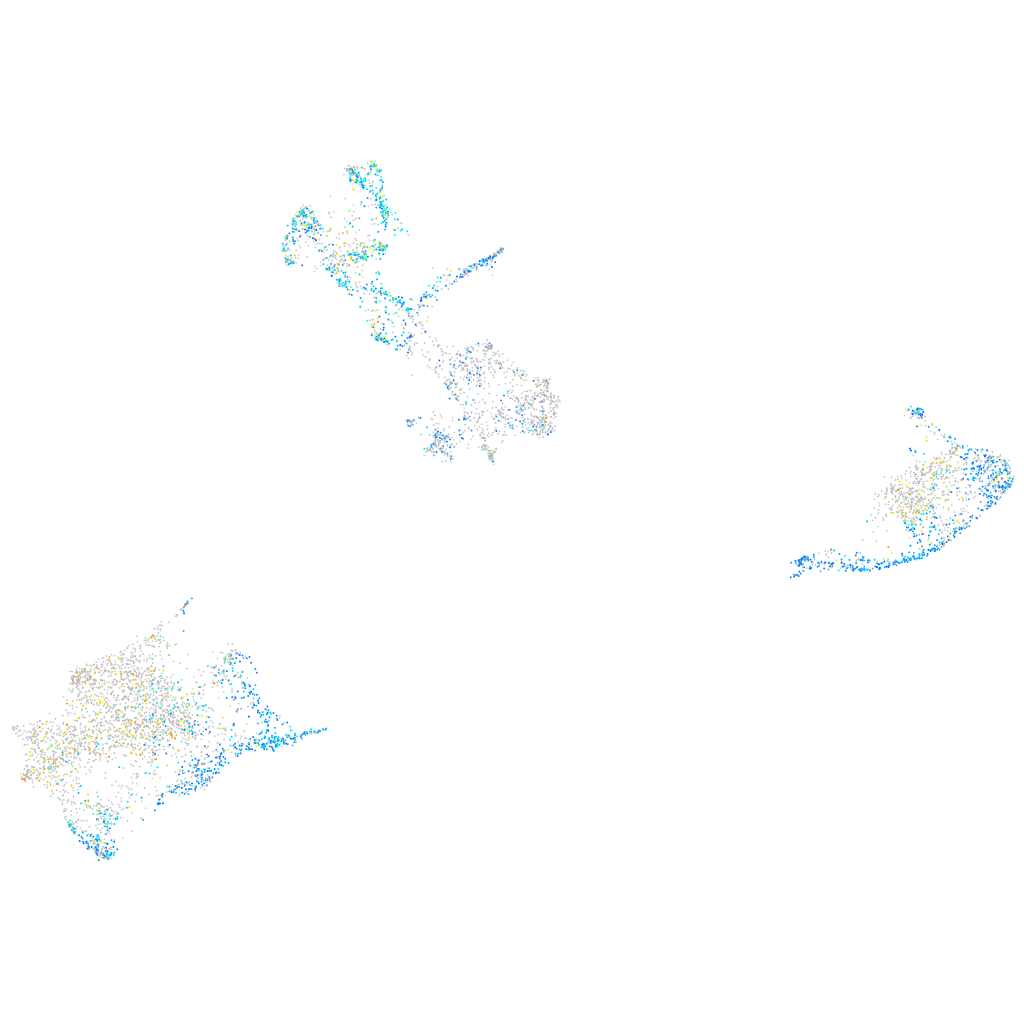
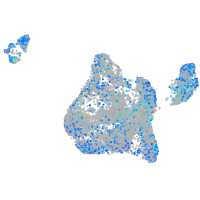
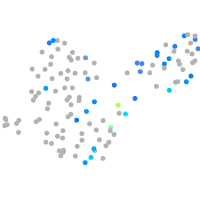
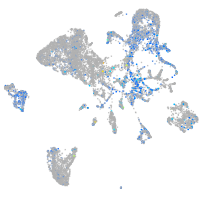
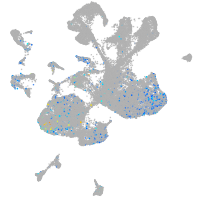
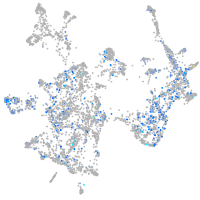
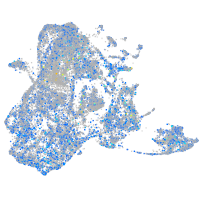
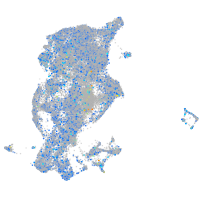
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| pnp4a | 0.283 | tuba1c | -0.171 |
| si:ch211-243a20.3 | 0.280 | gpm6aa | -0.154 |
| nme4 | 0.272 | CABZ01021592.1 | -0.144 |
| zgc:110239 | 0.270 | elavl3 | -0.144 |
| si:dkey-197i20.6 | 0.265 | stmn1b | -0.142 |
| agtrap | 0.256 | nova2 | -0.142 |
| zgc:110789 | 0.253 | tmsb | -0.140 |
| apoda.1 | 0.251 | hbbe1.3 | -0.139 |
| tfec | 0.249 | hbae3 | -0.136 |
| psph | 0.249 | hbae1.1 | -0.132 |
| guk1a | 0.248 | ptmaa | -0.129 |
| emp3b | 0.246 | si:dkey-251i10.2 | -0.126 |
| gpnmb | 0.241 | epb41a | -0.121 |
| alx4b | 0.240 | marcksl1b | -0.121 |
| alx4a | 0.239 | sncb | -0.120 |
| pltp | 0.237 | si:ch211-222l21.1 | -0.118 |
| phgdh | 0.236 | gch2 | -0.118 |
| trpm1b | 0.236 | elavl4 | -0.117 |
| si:ch73-89b15.3 | 0.234 | hbbe1.1 | -0.117 |
| smim29 | 0.234 | gng3 | -0.117 |
| si:ch211-256m1.8 | 0.231 | CU467822.1 | -0.116 |
| defbl1 | 0.230 | pvalb1 | -0.113 |
| BX571715.1 | 0.229 | marcksl1a | -0.111 |
| psat1 | 0.229 | fabp7a | -0.110 |
| tcirg1a | 0.227 | cadm3 | -0.107 |
| ltk | 0.227 | gng2 | -0.105 |
| akap12a | 0.224 | krt4 | -0.103 |
| unm-sa821 | 0.224 | tox | -0.102 |
| rnaset2 | 0.224 | zc4h2 | -0.100 |
| ctsba | 0.223 | hmgb3a | -0.098 |
| atox1 | 0.223 | actc1b | -0.098 |
| sytl2b | 0.221 | zgc:153426 | -0.097 |
| rgs3a | 0.221 | sox11a | -0.096 |
| tpd52l1 | 0.220 | vamp2 | -0.096 |
| lypc | 0.219 | stmn2a | -0.094 |